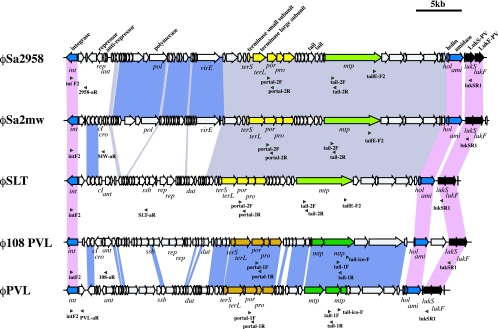
FIG. 3.
Structural comparison of 5 PVL phages. Structures of φ108PVL, φPVL, φSLT, φSa2mw, and φSa2958 are illustrated on the bases of the nucleotide sequences as follows: φ108PVL (DDBJ/EMBL/GenBank databases under accession no. AB243556), φPVL (DDBJ/EMBL/GenBank accession no. AB009866), φSLT (DDBJ/EMBL/GenBank accession no. NC_002661), φSa2mw (DDBJ/EMBL/GenBank accession no. BA000033), and φSa2958 (DDBJ/EMBL/GenBank accession no. AP009363). Genes having nucleotide identities of more than 90% are linked by blocks filled with colors as follows: pink, genes conserved among five PVL phages; light purple, genes conserved among φSa2958, φSa2mw, and φSLT; blue, genes conserved between φPVL and φ108PVL. The positions of primers for M-PCR 1 to 7 are indicated by green arrowheads. Blue arrows indicate ORFs well conserved in all PVL phages other than lukS and lukF genes; black arrows indicate lukS and lukF genes; light green and dark green arrows indicate genes encoding major tail proteins of the elongated-head type and the icosahedral-head type, respectively; and yellow arrows and ocher arrows indicate genes encoding terminase subunits (large and small), portal proteins, prohead proteases, and capsid proteins of the elongated-head type and the icosahedral-head type, respectively.