Abstract
Neisseria meningitidis strains (meningococci) with decreased susceptibility to penicillin (MICs, >0.06 μg/ml) have been reported in several parts of the world, but the prevalence of such isolates in Africa is poorly described. Data from an active national laboratory-based surveillance program from January 2001 through December 2005 were analyzed. A total of 1,897 cases of invasive meningococcal disease were reported, with an average annual incidence of 0.83/100,000 population. Of these cases, 1,381 (73%) had viable isolates available for further testing; 87 (6%) of these isolates tested intermediately resistant to penicillin (Peni). Peni meningococcal isolates were distributed throughout all provinces and age groups, and there was no association with outcome or human immunodeficiency virus infection. The prevalence of Peni was lower in serogroup A (7/295; 2%) than in serogroup B (24/314; 8%), serogroup C (9/117; 8%), serogroup Y (22/248; 9%), or serogroup W135 (25/396; 6%) (P = 0.02). Pulsed-field gel electrophoresis grouped 63/82 Peni isolates into nine clusters, mostly according to serogroup. The clustering of patterns from Peni isolates was not different from that of penicillin-susceptible isolates. Twelve sequence types were identified among 18 isolates arbitrarily selected for multilocus sequence typing. DNA sequence analysis of the penA gene identified 26 different alleles among the Peni isolates. Intermediate penicillin resistance is thus widespread among meningococcal serogroups, has been selected in a variety of lineages, and, to date, does not appear to be associated with increased mortality. This is the first report describing the prevalence and molecular epidemiology of Peni meningococcal isolates from sub-Saharan Africa.
Treatment of meningococcal disease with antibiotics has greatly reduced mortality, from almost 100% to less than 10% (50). Penicillin is used effectively in the treatment of meningococcal infections; however, strains with decreased susceptibility to penicillin (MICs, >0.06 μg/ml) have been reported in several countries (8, 11, 20, 44, 49), and some studies have shown that decreased susceptibility has been linked to a single clone (13, 38). The clinical significance of this increased resistance is unknown, and infections with these strains appear to respond to penicillin therapy (44).
Neisseria meningitidis contains three penicillin-binding proteins (PBPs): PBP1, encoded by ponA; PBP2, encoded by penA; and PBP3, encoded by pbp3 (2, 6, 19). In meningococci, penicillin nonsusceptibility is due to altered PBP2, causing intermediate resistance (MICs, 0.12 to 0.25 μg/ml) (4, 36), or, in rare cases, to the production of a plasmid-encoded β-lactamase, causing high-level resistance (MICs, >0.25 μg/ml) (9, 46).
Non-β-lactamase-producing penicillin-resistant meningococcal strains were first identified in Spain in 1985 (44). Subsequently, penicillin nonsusceptibility has been reported from several countries, although the frequency with which such isolates have been found varies widely. Surveillance studies of invasive isolates in the United Kingdom, Europe, and North America have reported prevalence rates for penicillin-nonsusceptible isolates ranging from 4% in the United States to 8% in Scotland, 23% in Sweden, and 30% and 38% in France and Portugal, respectively (3, 13, 25, 30, 35, 43). Seventeen percent of invasive isolates and 61% of carriage isolates showed decreased penicillin susceptibility in a study in Turkey (33).
Few studies have documented the existence of penicillin nonsusceptibility in Africa. A study in Casablanca, Morocco, reported an average rate of 4% for invasive meningococcal isolates intermediately resistant to penicillin from 1992 to 2000 (51), and strains from a meningitis outbreak in Ghana showed no evidence of resistance to penicillin (21). In 1987, β-lactamase-producing penicillin-resistant meningococcal isolates (MICs, >256 μg/ml) from two patients in South Africa were reported, but their mechanisms of resistance were never confirmed genotypically (9).
The aims of this study were to detect the prevalence of penicillin-nonsusceptible strains causing invasive disease in South Africa, to determine the clonality of these strains, and to examine the genetic diversity of their penA genes.
MATERIALS AND METHODS
National meningococcal surveillance.
Isolates were collected as part of a national laboratory-based surveillance system for invasive meningococcal disease in South Africa (24). Isolates and basic demographic data sent from approximately 120 laboratories throughout South Africa to the National Institute for Communicable Diseases in Johannesburg during the period from January 2001 through December 2005 were analyzed. A case of meningococcal disease was defined as isolation of Neisseria meningitidis from a cerebrospinal fluid (CSF) specimen or another specimen from a normally sterile site (e.g., blood or joint fluid). In 2003, the case definition was expanded to include cases in which specimens tested positive by latex agglutination, supported by Gram stain microscopy and/or PCR (34). In addition, expanded clinical and demographic information on patients at 15 sentinel hospitals in seven of nine provinces was collected. Newly captured data included outcome and human immunodeficiency virus (HIV) serological status.
Phenotypic strain characterization.
Bacteria were identified according to standardized procedures (48). Penicillin MICs for all viable isolates were determined by using the Etest (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar supplemented with 5% sheep blood and incubating at 37°C under 5% CO2. Broth microdilution MICs were determined for those isolates for which the penicillin MICs by the Etest were ≥0.25 μg/ml. Results were interpreted using the following MIC breakpoints, recommended by the Clinical and Laboratory Standards Institute (CLSI) performance standards: susceptibility to penicillin (Pens), ≤0.06 μg/ml; intermediate resistance (Peni), >0.06 to 0.25 μg/ml; resistance (Penr), ≥0.5 μg/ml (15). Nitrocefin (Oxoid Ltd., Basingstoke, England) was used to test all submitted isolates for β-lactamase production. Ceftriaxone, chloramphenicol, and rifampin were tested using disk diffusion screening, and MICs for all resistant strains were determined using Etest methodology (15, 16). Screening for fluoroquinolone nonsusceptibility, using ofloxacin discs and the zone size breakpoints stipulated for Neisseria gonorrhoeae (32), was initiated in 2003. In 2005, ciprofloxacin Etest methodology was introduced, and results were interpreted using the new MIC breakpoints provided by the CLSI (15). Serogroups were determined by using slide agglutination with polyclonal antisera to capsular polysaccharides A, C, X, Y, Z, and W135 and a monoclonal antiserum to capsular polysaccharide B (Remel Biotech Limited, Dartford, England). PCR was used to confirm serogrouping for all Peni isolates (39).
Genotypic strain characterization.
Isolates that were defined by their MICs (>0.06 μg/ml) as not susceptible to penicillin were selected for further characterization.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was carried out using the NheI restriction enzyme according to a previously described method (23), and restriction profiles were analyzed using GelCompar software (version 4.1; Applied Maths, Kortrijk, Belgium). Dendrograms were created using the unweighted-pair group method with arithmetic averages. The banding patterns were analyzed with the Dice coefficient. Optimization of 1.5% and a position tolerance of 1% were used for the band migration distance. PFGE profiles were visually inspected, and clusters were designated based on differences of ≤3 bands (42). This was congruent with a similarity index of approximately 80%. A PFGE cluster was therefore defined as three or more isolates sharing ≥80% similarity on the dendrogram. An arbitrary number was assigned to each cluster. The patterns of Peni isolates were compared to existing patterns for Pens meningococcal isolates stored in the database from previous studies (17, 47).
MLST.
Multilocus sequence typing (MLST) was performed on 18 isolates arbitrarily selected from each of the PFGE clusters and unrelated isolates. Sequence types (STs) were determined through the MLST website (http://pubmlst.org/neisseria) (27).
Restriction fragment length polymorphism (RFLP) of penA.
A 511-bp fragment of the transpeptidase region (3′ end) of the penA gene was amplified by PCR using previously described primers AA-1 and 99-2 (1). PCR products were subjected to TaqI restriction endonuclease digestion and electrophoresed through 2% agarose gels in order to determine the efficiency of this method in accurately differentiating Pens from Peni or Penr isolates (41). Eight penicillin-susceptible clinical isolates were included as controls (MICs, ≤0.06 μg/ml).
DNA sequencing of penA.
penA gene polymorphisms were determined by sequencing a 511-bp fragment of the transpeptidase region (3′ end) of the penA gene. Amplified products were purified using the QIAquick PCR purification kit (Qiagen Inc., Valencia, CA) and were sequenced using BigDye Terminator chemistry (version 3.1; Applied Biosystems, Foster City, CA) and an ABI 3130 capillary sequencer. Sequence analysis and alignment were carried out using DNAStar Lasergene software (version 7). penA alleles were assigned allele numbers based on the nomenclature described by Taha et al. (40) by comparing sequences with those currently available in the penA database (http://neisseria.org/perl/agdbnet/agdbnet.pl?file=penA.xml).
Statistical analysis.
Univariate analysis was performed using Fisher's exact test or the Mantel-Haenszel test for comparison of categorical variables. Analysis was performed with Epi Info software, version 6.04d (18), and Stata, version 9 (StataCorp Limited). Two-sided P values of <0.05 were considered significant throughout.
RESULTS
Meningococcal surveillance, 2001 to 2005.
For the period from January 2001 through December 2005, 1,897 cases of invasive meningococcal disease were reported, with an average annual incidence of 0.83/100,000 population (range, 0.59 to 1.16). Of all the cases identified in the 5-year period, 73% (1,381/1,897) had viable isolates. Age was known in 1,750 of 1,897 cases (92%). In 2005, the annual incidence was 1.16/100,000, and the highest age-specific incidence was in infants less than 1 year old (9.6/100,000 population). For the 5-year period, 67% (1,262/1,897) of cases were diagnosed from CSF culture specimens alone, 12% (229/1,897) from both CSF and blood culture specimens, and 21% (402/1,897) from blood culture specimens alone. Four cases were identified from synovial fluid culture specimens. Most cases of disease were reported from two provinces; Gauteng Province (55%; 1,049/1,897) and Western Cape Province (20%; 386/1,897). During this 5-year period, Western Cape Province reported a decreasing incidence of disease, predominantly due to serogroup B (47). In Gauteng Province, serogroup A disease with a stable incidence was predominant until 2004, when serogroup W135 became more prevalent, causing the majority of disease in 2005, with increased incidence (47). The percentages of cases due to serogroups C and Y, causing an average of 8% (117/1,381) and 18% (248/1,381) of all disease, respectively (17, 47), remained stable during the 5-year period.
During the period analyzed, 6% (87/1,381) of isolates were intermediately resistant to penicillin, with MICs ranging from 0.094 to 0.25 μg/ml (the MIC at which 50% of Peni isolates were inhibited was 0.125 μg/ml). No isolates tested fully resistant to penicillin or tested positive for β-lactamase production. Annual prevalences for the Peni isolates were 6% (14/231), 4% (8/192), 13% (33/264), 7% (20/281), and 3% (12/413) for 2001 to 2005, respectively. All Peni isolates were susceptible to chloramphenicol, ceftriaxone, and rifampin. Similarly, all Pens isolates were susceptible to chloramphenicol and ceftriaxone, and 0.3% (4/1,293) were not susceptible to rifampin. All isolates were susceptible to fluoroquinolones.
Invasive meningococcal disease caused by Peni strains did not differ from that caused by Pens strains with regard to the percentage of cases in which patients were children less than 5 years old (50% [42/84] of Peni cases versus 44% [526/1,200] of Pens cases; P = 0.27) or male (52% [44/84] of Peni cases versus 58% [730/1,255] of Pens cases; P = 0.3). The prevalence of Peni was lower among cases in which N. meningitidis was isolated from CSF (5%; 59/1,075) than among those in which the organism was isolated from other sterile sites (9%; 28/306) (P = 0.02). During the period of enhanced surveillance (2003 to 2005), only five patients from whom Peni meningococci were isolated were tested for HIV; of these, 1 patient was HIV positive compared to 79/183 (43%) Pens patients. The in-hospital case fatality rate was 4/23 (17%) for Peni cases compared to 57/359 (16%) for Pens cases (P = 0.85).
The prevalence of Peni isolates for the 5-year period differed by serogroup, with serogroup A demonstrating the lowest prevalence (7/295; 2%); other prevalences were 8% (24/314) for serogroup B, 8% (9/117) for serogroup C, 9% (22/248) for serogroup Y, and 6% (25/396) for serogroup W135 (P = 0.02). Prevalence fluctuated within serogroups over the years; for example, Peni prevalences in serogroup W135 were 15% (2/13), 4% (1/24), 12% (3/26), 12% (9/77), and 4% (10/256) for 2001 to 2005, respectively. The prevalences of Peni in serogroup B ranged from 2% (1/58) in 2005 to 13% (10/76) in 2003. The Peni isolates were identified in all nine provinces of South Africa. The percentages of Peni isolates in the different provinces were as follows: 13% (7/52) in the Eastern Cape, 9% (9/99) in Free State, 5% (42/813) in Gauteng, 11% (5/46) in KwaZulu-Natal, 5% (1/19) in Limpopo, 5% (2/43) in Mpumalanga, 6% (1/16) in Northern Cape, 4% (2/51) in Northwest, and 7% (18/242) in the Western Cape. The relative percentages of Peni and Pens isolates did not differ between provinces (P = 0.23).
Molecular characterization.
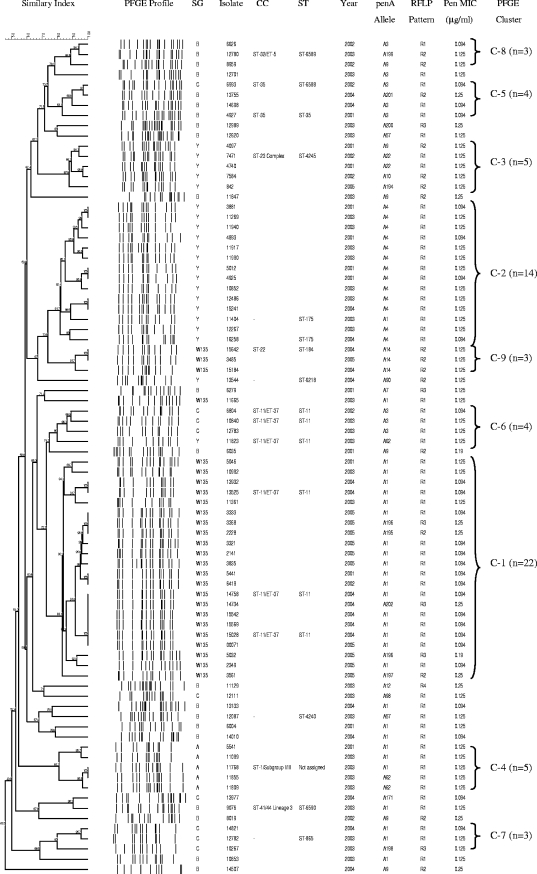
PFGE patterns were obtained for 82/87 Peni isolates and were grouped into 9 clusters and 19 unrelated isolates (Fig. 1). Cluster 1 (22/82; 27%) comprised W135 strains isolated mostly during 2004 and 2005, all of which also belonged to a larger Pens PFGE cluster representing 93% of all W135 isolates collected nationally during 2000 to 2005 (47). Cluster 2 (14/82; 17%) comprised isolates belonging to serogroup Y. The serogroup Y isolates in this cluster were related by PFGE to a larger clone including serogroup Y Pens isolates, analyzed for this time period. Cluster 3 comprised five serogroup Y isolates (6%). Cluster 4 consisted of five serogroup A isolates, all of which belonged to a larger clone, including Pens isolates, originating from Gauteng Province. Cluster 5 (4/82; 5%) comprised serogroup B and C isolates. Cluster 6 consisted of three serogroup C isolates and one serogroup Y isolate (4/82; 5%). The remaining three clusters were made up of three isolates each, which belonged to serogroups C, B, and W135, respectively. Nineteen isolates were unrelated to the nine clusters and belonged to serogroups B (n = 15), C (n = 2), Y (n = 1), and W135 (n = 1). When analyzed together with Pens isolates, serogroup B and C Peni isolates were found to be heterogeneous and to belong to several different clusters (data not shown).
FIG. 1.
Dendrogram representing the PFGE patterns of 82 N. meningitidis isolates intermediately resistant to penicillin that caused invasive disease in South Africa in 2001 to 2005. Similarity values are given on the dendrogram branches. SG, serogroup; CC, clonal complex.
MLST identified 12 STs among 18 arbitrarily selected Peni isolates, including 3 novel STs (Fig. 1). Three isolates from cluster 1, representing serogroup W135, were identified as ST-11 of the ST-11/ET-37 clonal complex, together with two serogroup C isolates and one serogroup Y isolate from cluster 6 that were 75% related to the cluster 1 isolates. Two isolates from cluster 2 were ST-175 (serogroup Y). One isolate from cluster 3 was ST-4245 of the ST-23 complex (serogroup Y). A serogroup A isolate from cluster 4 belonged to the ST-1/subgroup I/II clonal complex. No ST was assigned to this isolate, because sequence data were available for only six of seven alleles (repeated attempts to sequence the pdhC allele failed). Two serogroup B isolates from cluster 5 were ST-35 and a new ST, ST-6588, respectively; both of these belong to the ST-35 complex and share identical alleles at five of seven loci. The remaining STs were ST-184 (serogroup W135), ST-6218 (serogroup Y), the new ST-6589 (serogroup B), the new ST-6590 (serogroup B), ST-4240 (serogroup B), and ST-865 (serogroup C). Eleven STs were assigned to 38 of 1,294 Pens isolates; of these, six STs were also observed among the Peni isolates (17, 47).
RFLP of the penA gene for 87 Peni isolates revealed four restriction endonuclease patterns (Fig. 1). All isolates for which the MIC was ≤0.094 μg/ml (n = 27) had the same pattern (wild type, or RFLP-1). Forty-eight isolates for which the MIC was 0.125 μg/ml displayed either the RFLP-1 (n = 35) or the RFLP-2 or -3 (n = 13) pattern. Isolates for which the MIC was ≥0.19 μg/ml (n = 12) had altered patterns (RFLP-2, -3, or -4). Eight penicillin-susceptible control isolates all displayed the wild-type pattern.
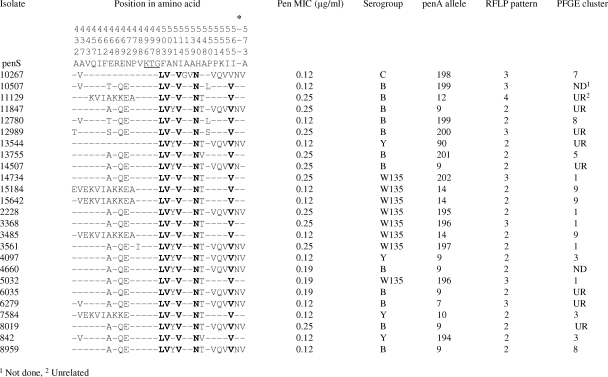
DNA sequence analysis of the transpeptidase region of the penA gene from 87 Peni isolates identified 24 different alleles by using the online database, including 9 novel penA alleles that were not listed in the database and were assigned new allele types (Fig. 1 and 2). Amino acid substitutions were observed in PBP2 of only 25/87 (29%) Peni isolates. Twelve isolates for which the MIC was equal to 0.125 μg/ml and all isolates for which the MIC was greater than 0.125 μg/ml (n = 13) displayed altered penA alleles with 10 to 15% nucleotide variation in the transpeptidase region, resulting in 9 to 17 amino acid substitutions in PBP2 (Fig. 2). Eight amino acid substitutions were common to 23 of the 25 isolates; the remaining 2 isolates (isolates 10267 and 13544) harbored six of the eight common substitutions. Twelve isolates harbored an additional Asn residue inserted between residues Thr572 and Ala573 at the 3′ end of the fragment.
FIG. 2.
Amino acid alignment showing substitutions in the transpeptidase region of PBP2 for 25 N. meningitidis isolates from South Africa, 2001 to 2005, with intermediate resistance to penicillin. Dashes indicate residues identical to those in the sequence of the Pens strain. The conserved KTG motif in the Pens strain is underlined. Boldfaced residues are common to all 25 isolates. An asterisk marks the position where some isolates have an Asn insertion.
DISCUSSION
Our data show that the prevalence of isolates with decreased susceptibility to penicillin is lower in South Africa than in Europe (3, 13) but similar to the prevalences reported for the United Kingdom (30) and the United States (35). Although there were fluctuations, the prevalence exceeded 10% only in 1 of 5 years studied, and no increase in prevalence was observed over the study period. Some studies have shown a steady increase in the prevalence of Peni isolates since they were first detected (11, 36), whereas others have shown a stable prevalence over time (28, 30, 51). We found that serogroup A was associated with the lowest prevalence of decreased penicillin susceptibility. Other studies have shown the prevalence of Peni isolates to be higher among serogroup C and W135 isolates (3, 11, 36).
Overall, the Peni isolates analyzed for this period did not differ from the Pens isolates with regard to patient characteristics such as age or gender, and there was no association with outcome or HIV infection. During the study period evaluated for outcome (2003 to 2005), extended-spectrum cephalosporins were available in clinics and hospitals for empirical treatment of acute bacterial meningitis and were recommended for both children and adults (31); this may have influenced the lack of association with outcome that we observed. In our study, Peni isolates were more likely to be isolated from blood culture specimens than from CSF specimens. Oral antibiotics given to ill patients prior to admission may affect bacteremic organisms, rendering blood sterile, with less impact on organisms in the CSF. Our sample of bacteremic isolates may therefore be biased toward the more resistant strains.
PFGE showed a high degree of genetic diversity among the South African Peni isolates, with no dominant clone. Isolates clustered mostly according to serogroup. Previous work has shown that serogroup B and C isolates are genetically diverse in South Africa (17), and similarly, the Peni isolates belonging to these serogroups were genetically heterogeneous. In contrast, serogroup A, W135, and Y isolates are highly clonal in South Africa (17), and the Peni isolates belonging to these serogroups mimicked this trend. Several STs were identified among the Peni isolates, some of which have been described in Pens isolates (17, 47). Several studies have, like ours, found Peni strains to be genetically diverse (3, 25, 36). However, some studies have reported the emergence and spread of virulent clones intermediately resistant to penicillin (7, 12, 38). A Peni serogroup C clone caused an outbreak in Saskatchewan, Canada, in 1993 (7). A surveillance study in Portugal in 2000 to 2001 showed that penicillin resistance was significantly associated with serogroup C isolates displaying the C:2b:P1.2 phenotype (12). In Italy, an increase in serogroup C disease occurred in 2000 to 2003 (38). The majority of isolates (42%) belonged to a penicillin-nonsusceptible serogroup C clone (C:2b:P1.5) similar to that found in Portugal.
Some studies have suggested that restriction digestion of the penA gene is a rapid and reliable method for screening for penicillin nonsusceptibility in cultures (1, 29) and may also be useful for determining penicillin nonsusceptibility in culture-negative cases of meningococcal infection. However, our data show that only those Peni isolates that harbored penA alleles with nonsynonymous mutations displayed restriction profiles different from that of the wild type. Several Peni isolates displayed restriction patterns identical to those for Pens isolates, indicating that they would have been misclassified as Pens by use of this screening method.
penA sequences from non-penicillin-susceptible strains have been shown to be variable and highly divergent from those of wild-type strains, particularly in the 3′ transpeptidase-encoding region (2). A MIC of >0.06 μg/ml has been established as the breakpoint for defining intermediate resistance to penicillin by use of broth microdilution (15). Our data show that all isolates for which the Etest MIC was ≥0.19 μg/ml had polymorphic penA alleles. The majority of isolates for which the Etest MIC was 0.125 μg/ml had penA alleles that were almost identical to the wild-type allele, differing by only a few nucleotide substitutions. Other studies have shown that a MIC of ≥0.125 μg/ml was associated with polymorphic penA genes (29, 43). Arreaza et al. have suggested that a MIC of 0.094 μg/ml should be used to define the intermediate breakpoint for penicillin (5). All of the South African isolates for which the MIC was ≤0.094 μg/ml had wild-type penA alleles. Phenotypic antibiotic susceptibility testing is, however, difficult to compare between laboratories in different countries because of differences in reagents and methodology (45).
The alteration of only one PBP suggests that penicillin resistance in N. meningitidis is still evolving, and the accumulation of additional altered PBPs may confer a higher level of resistance. Nucleotide sequencing of the penA gene showed a high degree of sequence diversity among the Peni isolates in this study, suggesting that these genes have arisen independently. The majority of Peni isolates harbored penA alleles with synonymous nucleotide substitutions, resulting in no amino acid alterations in PBP2. Approximately one-third of the Peni isolates had an altered PBP2. Eight amino acid changes were common to all isolates that had an altered penA allele (except for two isolates that had only six of these substitutions). The eight substitutions were the same as those previously described by Antignac et al. (2) and have been shown to correlate with the same region in Streptococcus pneumoniae PBP2X, which forms part of the active site for β-lactam binding. These investigators also showed that a PCR-amplified fragment harboring these eight mutations was sufficient to confer resistance on a penicillin-susceptible strain. Transformation studies with N. gonorrhoeae have shown that the presence of an additional aspartic acid residue in PBP2 results in a decreased affinity for penicillin; however, the presence of any other additional amino acids did not confer resistance (10). The significance of the Asn insertion in PBP2 of some of the South African N. meningitidis isolates is unclear, since it was present only in some altered penA alleles.
Nonsusceptibility to rifampin, chloramphenicol, and ceftriaxone has been reported previously but remains rare, with studies reporting either no reduced susceptibility or low prevalences (22, 26, 37, 51). Similarly, we report no reduced susceptibility to chloramphenicol or ceftriaxone and an extremely low prevalence of nonsusceptibility to rifampin. In 2007 to 2008, three cases of infection with epidemiologically and genotypically linked fluoroquinolone-resistant serogroup B meningococci were reported from the North Dakota-Minnesota region of the United States (14). The use of ciprofloxacin for chemoprophylaxis was subsequently suspended in this region, until further notice. We report no fluoroquinolone resistance among our meningococcal isolates.
To our knowledge, this is the first publication describing meningococci from sub-Saharan Africa that are intermediately resistant to penicillin. These isolates do not, at this time, appear to be due to the spread of a single clone. Continued surveillance, in South Africa and elsewhere in Africa, is necessary for monitoring ongoing trends in antimicrobial susceptibility, in particular the emergence of resistant strains that may pose therapeutic problems.
Acknowledgments
We thank all laboratory and clinical staff throughout South Africa for contributing to national surveillance; Garry Coulson, Olga Hattingh, Lenny Lengwati, Ruth Mpembe, Thomas Rafundisani, and Happy Skosana for technical assistance; Muzi Hlanzi and Ethel Maringa for data management; and Leonard Mayer for kind review of the manuscript.
We also thank the following members of GERMS-SA: Sandeep Vasaikar (University of Transkei, Mthatha, Eastern Cape); Nolan Janse van Rensberg and Peter Smith (University of the Free State, Bloemfontein, Free State); Khatija Ahmed, Anwar Hoosen, Ruth Lekalakala, and Pyu Pyu Sein (University of Limpopo—Medunsa Campus, Garankuwa, Gauteng); Heather Crewe-Brown, Charles Feldman, Alan Karstaedt, Olga Perovic, and Jeannette Wadula (University of the Witwatersrand, Johannesburg, Gauteng); Mike Dove, Kathy Lindeque, Linda Meyer, and Gerhard Weldhagen (University of Pretoria, Pretoria, Gauteng); Wim Sturm and Trusha Vanmali (University of KwaZulu-Natal, Durban, KwaZulu-Natal); Ken Hamese (Polokwane/Mankweng Hospital Complex, Polokwane, Limpopo); Keith Bauer, Greta Hoyland, and Charles Mutanda (National Health Laboratory Service, Mpumalanga); Rena Hoffmann and Lynne Liebowitz (University of Stellenbosch, Stellenbosch, Western Cape); Denise Roditi, John Simpson, and Andrew Whitelaw (University of Cape Town, Cape Town, Western Cape); Adrian Brink (AMPATH Laboratories, Johannesburg, Gauteng); Claire Heney (Lancet Laboratories, Johannesburg, Gauteng); Marthinus Senekal (PathCare Laboratories, Cape Town, Western Cape); and John Frean, Karen Keddy, Kerrigan McCarthy, Elizabeth Prentice, Vanessa Quan, and Koshika Soma (National Institute for Communicable Diseases, Johannesburg, Gauteng).
This study was funded by the Medical Research Council, the National Institute for Communicable Diseases (NICD) of the National Health Laboratory Service (NHLS), and the University of the Witwatersrand, South Africa, and by funds from the U.S. Agency for International Development's Antimicrobial Resistance Initiative, transferred via a cooperative agreement (U60/CCU022088) from the Centers for Disease Control and Prevention (CDC), Atlanta, GA. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the CDC.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Antignac, A., J. M. Alonso, and M. K. Taha. 2001. Nonculture prediction of Neisseria meningitidis susceptibility to penicillin. Antimicrob. Agents Chemother. 453625-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antignac, A., I. G. Boneca, J. C. Rousselle, A. Namane, J. P. Carlier, J. A. Vazquez, A. Fox, J. M. Alonso, and M. K. Taha. 2003. Correlation between alterations of the penicillin-binding protein 2 and modifications of the peptidoglycan structure in Neisseria meningitidis with reduced susceptibility to penicillin G. J. Biol. Chem. 27831529-31535. [DOI] [PubMed] [Google Scholar]
- 3.Antignac, A., M. Ducos-Galand, A. Guiyoule, R. Pires, J. M. Alonso, and M. K. Taha. 2003. Neisseria meningitidis strains isolated from invasive infections in France (1999-2002): phenotypes and antibiotic susceptibility patterns. Clin. Infect. Dis. 37912-920. [DOI] [PubMed] [Google Scholar]
- 4.Antignac, A., P. Kriz, G. Tzanakaki, J. M. Alonso, and M. K. Taha. 2001. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J. Antimicrob. Chemother. 47285-296. [DOI] [PubMed] [Google Scholar]
- 5.Arreaza, L., C. Salcedo, B. Alcala, M. J. Uria, R. Abad, R. Enriquez, and J. A. Vazquez. 2004. Sequencing of Neisseria meningitidis penA gene: the key to success in defining penicillin G breakpoints. Antimicrob. Agents Chemother. 48358-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour, A. G. 1981. Properties of penicillin-binding proteins in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 19316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blondeau, J. M., F. E. Ashton, M. Isaacson, Y. Yaschuck, C. Anderson, and G. Ducasse. 1995. Neisseria meningitidis with decreased susceptibility to penicillin in Saskatchewan, Canada. J. Clin. Microbiol. 331784-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boras, A., D. Bozinovic, F. C. Tenover, and T. Popovic. 2001. First report of Neisseria meningitidis intermediately resistant to penicillin in Croatia. J. Clin. Microbiol. 39823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botha, P. 1988. Penicillin-resistant Neisseria meningitidis in southern Africa. Lancet i54. [DOI] [PubMed] [Google Scholar]
- 10.Brannigan, J. A., I. A. Tirodimos, Q. Y. Zhang, C. G. Dowson, and B. G. Spratt. 1990. Insertion of an extra amino acid is the main cause of the low affinity of penicillin-binding protein 2 in penicillin-resistant strains of Neisseria gonorrhoeae. Mol. Microbiol. 4913-919. [DOI] [PubMed] [Google Scholar]
- 11.Brown, S., G. Riley, and F. Jamieson. 2001. Neisseria meningitidis with decreased susceptibility to penicillin in Ontario, Canada 1997-2000. Can. Commun. Dis. Rep. 2773-75. [PubMed] [Google Scholar]
- 12.Caniça, M., R. Dias, and E. Ferreira. 2004. Neisseria meningitidis C:2b:P1.2,5 with intermediate resistance to penicillin, Portugal. Emerg. Infect. Dis. 10526-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caniça, M., R. Dias, B. Nunes, L. Carvalho, and E. Ferreira. 2004. Invasive culture-confirmed Neisseria meningitidis in Portugal: evaluation of serogroups in relation to different variables and antimicrobial susceptibility (2000-2001). J. Med. Microbiol. 53921-925. [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 2008. Emergence of fluoroquinolone-resistant Neisseria meningitidis—Minnesota and North Dakota, 2007-2008. MMWR Morb. Mortal. Wkly. Rep. 57173-175. [PubMed] [Google Scholar]
- 15.CLSI. 2005. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. CLSI document M100-S, 15th ed. CLSI, Wayne, PA.
- 16.CLSI. 2006. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. CLSI document M100-S, 16th ed. CLSI, Wayne, PA.
- 17.Coulson, G. B., A. von Gottberg, M. du Plessis, A. M. Smith, L. de Gouveia, and K. P. Klugman. 2007. Meningococcal disease in South Africa, 1999-2002. Emerg. Infect. Dis. 13273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean, A. D., J. A. Dean, and J. H. Burton. 1996. EpiInfo v6.04: a word processing, database and statistics program for epidemiology on microcomputers. Centers for Disease Control and Prevention, Atlanta, GA.
- 19.Dougherty, T. J., A. E. Koller, and A. Tomasz. 1981. Competition of beta-lactam antibiotics for the penicillin-binding proteins of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 20109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira, E., and M. Caniça. 2002. Invasive meningococci with reduced susceptibility to penicillin in Portugal. J. Antimicrob. Chemother. 49424-425. [DOI] [PubMed] [Google Scholar]
- 21.Gagneux, S., A. Hodgson, I. Ehrhard, G. Morelli, B. Genton, T. Smith, M. Tanner, F. Binka, M. Achtman, and G. Pluschke. 2000. Microheterogeneity of serogroup A (subgroup III) Neisseria meningitidis during an outbreak in northern Ghana. Trop. Med. Int. Health 5280-287. [PubMed] [Google Scholar]
- 22.Galimand, M., G. Gerbaud, M. Guibourdenche, J. Y. Riou, and P. Courvalin. 1998. High-level chloramphenicol resistance in Neisseria meningitidis. N. Engl. J. Med. 339868-874. [DOI] [PubMed] [Google Scholar]
- 23.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 352977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huebner, R. E., K. P. Klugman, U. Matai, R. Eggers, G. Hussey, et al. 1999. Laboratory surveillance for Haemophilus influenzae type B meningococcal, and pneumococcal disease. S. Afr. Med. J. 89924-925. [PubMed] [Google Scholar]
- 25.Jackson, L. A., F. C. Tenover, C. Baker, B. D. Plikaytis, M. W. Reeves, S. A. Stocker, R. E. Weaver, J. D. Wenger, et al. 1994. Prevalence of Neisseria meningitidis relatively resistant to penicillin in the United States, 1991. J. Infect. Dis. 169438-441. [DOI] [PubMed] [Google Scholar]
- 26.Jackson, L. A., and J. D. Wenger. 1993. Laboratory-based surveillance for meningococcal disease in selected areas, United States, 1989-1991. MMWR CDC Surveill. Summ. 4221-30. [PubMed] [Google Scholar]
- 27.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, D. M., and E. M. Sutcliffe. 1990. Meningococci with reduced susceptibility to penicillin. Lancet 335863-864. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen, J. H., S. A. Crawford, and K. R. Fiebelkorn. 2005. Susceptibility of Neisseria meningitidis to 16 antimicrobial agents and characterization of resistance mechanisms affecting some agents. J. Clin. Microbiol. 433162-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyaw, M. H., J. C. Bramley, S. Clark, P. Christie, I. G. Jones, and H. Campbell. 2002. Prevalence of moderate penicillin resistant invasive Neisseria meningitidis infection in Scotland, 1994-9. Epidemiol. Infect. 128149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Department of Health, South Africa. 2006. Standard treatment guidelines and essential drugs list for South Africa: hospital level, adults. http://www.doh.gov.za/docs/index.html.
- 32.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing; fourteenth informational supplement. NCCLS document M100-S, 14th ed. NCCLS, Wayne, PA.
- 33.Punar, M., H. Eraksoy, A. A. Cagatay, H. Ozsut, A. Kaygusuz, S. Calangu, and M. Dilmener. 2002. Neisseria meningitidis with decreased susceptibility to penicillin in Istanbul, Turkey. Scand. J. Infect. Dis. 3411-13. [DOI] [PubMed] [Google Scholar]
- 34.Rådstrom, P., A. Backman, N. Qian, P. Kragsbjerg, C. Pahlson, and P. Olcen. 1994. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J. Clin. Microbiol. 322738-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenstein, N. E., S. A. Stocker, T. Popovic, F. C. Tenover, B. A. Perkins, and the Active Bacterial Core Surveillance (ABCs) Team. 2000. Antimicrobial resistance of Neisseria meningitidis in the United States, 1997. Clin. Infect. Dis. 30212-213. [DOI] [PubMed] [Google Scholar]
- 36.Sáez-Nieto, J. A., R. Lujan, S. Berron, J. Campos, M. Vinas, C. Fuste, J. A. Vazquez, Q. Y. Zhang, L. D. Bowler, J. V. Martinez-Suarez, et al. 1992. Epidemiology and molecular basis of penicillin-resistant Neisseria meningitidis in Spain: a 5-year history (1985-1989). Clin. Infect. Dis. 14394-402. [DOI] [PubMed] [Google Scholar]
- 37.Stefanelli, P., C. Fazio, G. La Rosa, C. Marianelli, M. Muscillo, and P. Mastrantonio. 2001. Rifampicin-resistant meningococci causing invasive disease: detection of point mutations in the rpoB gene and molecular characterization of the strains. J. Antimicrob. Chemother. 47219-222. [DOI] [PubMed] [Google Scholar]
- 38.Stefanelli, P., C. Fazio, A. Neri, T. Sofia, and P. Mastrantonio. 2004. Emergence in Italy of a Neisseria meningitidis clone with decreased susceptibility to penicillin. Antimicrob. Agents Chemother. 483103-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taha, M. K. 2000. Simultaneous approach for nonculture PCR-based identification and serogroup prediction of Neisseria meningitidis. J. Clin. Microbiol. 38855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taha, M. K., J. A. Vazquez, E. Hong, D. E. Bennett, S. Bertrand, S. Bukovski, M. T. Cafferkey, F. Carion, J. J. Christensen, M. Diggle, G. Edwards, R. Enriquez, C. Fazio, M. Frosch, S. Heuberger, S. Hoffmann, K. A. Jolley, M. Kadlubowski, A. Kechrid, K. Kesanopoulos, P. Kriz, L. Lambertsen, I. Levenet, M. Musilek, M. Paragi, A. Saguer, A. Skoczynska, P. Stefanelli, S. Thulin, G. Tzanakaki, M. Unemo, U. Vogel, and M. L. Zarantonelli. 2007. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob. Agents Chemother. 512784-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taha, M. K., M. L. Zarantonelli, A. Neri, R. Enriquez, J. A. Vazquez, and P. Stefanelli. 2006. Interlaboratory comparison of PCR-based methods for detection of penicillin G susceptibility in Neisseria meningitidis. Antimicrob. Agents Chemother. 50887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thulin, S., P. Olcen, H. Fredlund, and M. Unemo. 2006. Total variation in the penA gene of Neisseria meningitidis: correlation between susceptibility to beta-lactam antibiotics and penA gene heterogeneity. Antimicrob. Agents Chemother. 503317-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Esso, D., D. Fontanals, S. Uriz, M. A. Morera, T. Juncosa, C. Latorre, and M. Duran. 1987. Neisseria meningitidis strains with decreased susceptibility to penicillin. Pediatr. Infect. Dis. J. 6438-439. [DOI] [PubMed] [Google Scholar]
- 45.Vázquez, J. A., L. Arreaza, C. Block, I. Ehrhard, S. J. Gray, S. Heuberger, S. Hoffmann, P. Kriz, P. Nicolas, P. Olcen, A. Skoczynska, L. Spanjaard, P. Stefanelli, M. K. Taha, and G. Tzanakaki. 2003. Interlaboratory comparison of agar dilution and Etest methods for determining the MICs of antibiotics used in management of Neisseria meningitidis infections. Antimicrob. Agents Chemother. 473430-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vázquez, J. A., A. M. Enriquez, L. de la Fuente, S. Berron, and M. Baquero. 1996. Isolation of a strain of beta-lactamase-producing Neisseria meningitidis in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 15181-182. [DOI] [PubMed] [Google Scholar]
- 47.von Gottberg, A., M. du Plessis, C. Cohen, E. Prentice, S. Schrag, L. de Gouveia, G. Coulson, G. de Jong, and K. Klugman. 2008. Emergence of endemic serogroup W135 meningococcal disease associated with a high mortality rate in South Africa. Clin. Infect. Dis. 46377-386. [DOI] [PubMed] [Google Scholar]
- 48.Winn, W. C. J., S. D. Allen, W. M. Janda, E. M. Koneman, G. W. Procop, P. C. Schreckenberger, and G. L. Woods. 2006. Koneman's color atlas and textbook of diagnostic microbiology, 6th ed., p. 566-622. Lippincott Williams & Wilkins, Philadelphia, PA.
- 49.Woods, C. R., A. L. Smith, B. L. Wasilauskas, J. Campos, and L. B. Givner. 1994. Invasive disease caused by Neisseria meningitidis relatively resistant to penicillin in North Carolina. J. Infect. Dis. 170453-456. [DOI] [PubMed] [Google Scholar]
- 50.World Health Organization. 2007. Control of epidemic meningococcal disease. WHO practical guidelines, 2nd ed. WHO/EMC/BAC/98.3. www.who.int/csr/resources/publications/meningitis/WHO_EMC_BAC_98_3_EN/en/.
- 51.Zerouali, K., N. Elmdaghri, M. Boudouma, and M. Benbachir. 2002. Serogroups, serotypes, serosubtypes and antimicrobial susceptibility of Neisseria meningitidis isolates in Casablanca, Morocco. Eur. J. Clin. Microbiol. Infect. Dis. 21483-485. [DOI] [PubMed] [Google Scholar]