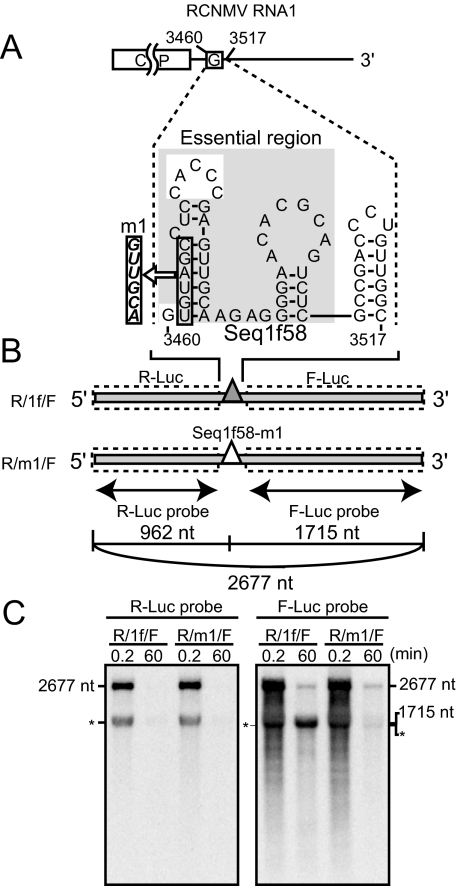
FIG. 4.
Seq1f58 is sufficient for the accumulation of its downstream nonviral RNA fragments. (A) Schematic representation of the essential nucleotide sequences required for the accumulation of SR1f (Seq1f58). The G's in the open boxes indicate the nucleotide residue at the 5′ end of SR1f. The nucleotide sequence in the gray shaded box indicates the essential nucleotide sequence required for the accumulation of SR1f. A secondary structure of Seq1f58 was predicted by using mfold (version 2.3) (50, 58) at 17°C. (B) Schematic representation of dicistronic mRNA. Seq1f58 was inserted between two different reporter ORFs, Renilla luciferase and firefly luciferase (R/1f/F). R/1f/F is 2,677 nt in length. The number of nucleotide residues from the 5′ end to just before the first guanine residue in the Seq1f58 is 962, and the number from the first guanine residue in the Seq1f58 to the 3′ end is 1,715. The filled and open triangles show Seq1f58 and Seq1f58 with the m1 mutation (Seq1f58-m1), respectively. The regions covered by RNA probes are shown as thin arrows. (C) Accumulation of degradation intermediates derived from R/1f/F or R/m1/F in BYL at 0.2 and 60 min after incubation. R/m1/F has the m1 mutation in Seq1f58. R/1f/F or R/m1/F (0.55 pmol) was incubated in 50 μl BYL at 17°C. Total RNA was extracted at 0.2 min and 60 min after incubation and used for Northern blotting with DIG-labeled RNA probes specific for the F-Luc ORF and R-Luc ORF. The sizes of the bands are indicated on the right or the left side. The asterisks indicate an unexpected band which was detected by both the R-Luc and F-Luc probes at 0.2 or 60 min after incubation.