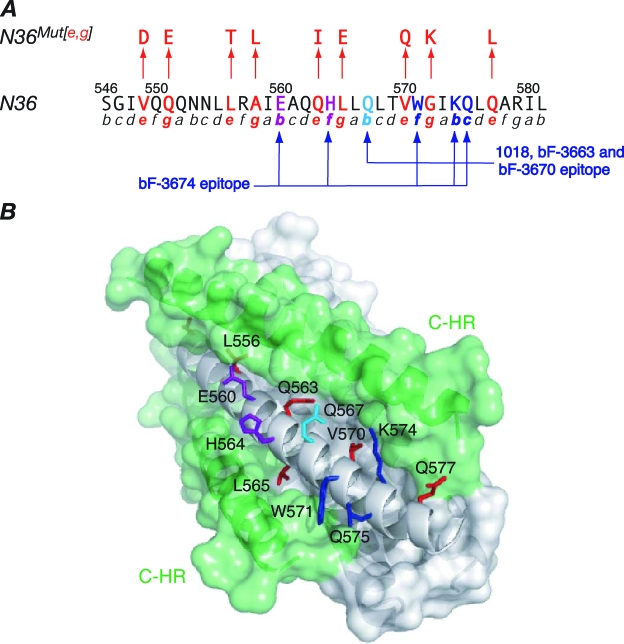
FIG. 2.
N36Mut(e,g) and the epitopes for the neutralizing (Fab 3674) and nonneutralizing (Fabs 3663, 3670, and 1018) MAbs directed against the N-HR of gp41. (A) Sequence of N36 (residues 546 to 581 of gp41), with the helical wheel positions in the N-HR trimer indicated below the sequence (5). Positions e and g (red), which contact the C-HR helix in the 6-HB conformation of gp41, are mutated in N36Mut(e,g) (2). As a result, N36Mut(e,g) forms a trimer that does not interact with C34 (a peptide comprising residues 628 to 661 of the C-HR of gp41) but can form heterotrimers with the N-HR of gp41, thereby inhibiting fusion (2). The epitopes of the neutralizing bF-3674 Fab (E560, H563, W571, K574, and Q575) and the nonneutralizing bF-1018, bF-3663, and bF-3670 Fabs (Q567, W571, K474, and Q575), previously delineated by alanine-scanning mutagenesis (17), are indicated. (B) Surface representation of the 6-HB core of HIV-1 gp41 (5, 49, 52). The trimeric N-HR helices are in gray, and the C-HR helices (two in the view shown) in green. Solvent-exposed N-HR residues that comprise the epitopes of the Fabs and lie in a shallow groove between two C-HR helices are shown as stick diagrams and colored as follows: residues common to the epitopes of both the neutralizing and nonneutralizing Fabs, blue; residues that are part of only the neutralizing Fab epitope, purple; the residue that is part of only the nonneutralizing Fab epitope, light blue. Residues colored in red are N-HR residues at positions e and g of the helical wheel that interact with the C-HR helices.