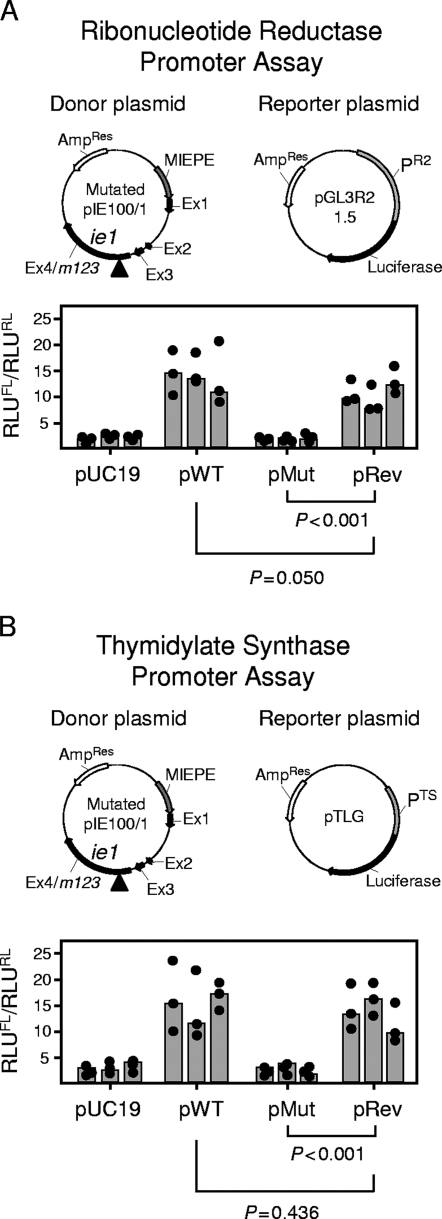
FIG. 2.
Impact of the mutation Y165C on the transactivation of cellular genes involved in dNTP biosynthesis. (A) Transactivation of the RNR promoter PR2. (B) Transactivation of the TS promoter PTS. DLR assays were performed with growth-arrested NIH 3T3 cells transiently cotransfected with an IE1-encoding donor plasmid, the respective firefly luciferase-encoding reporter plasmid, and the Renilla luciferase-encoding plasmid pRL-TK for standardization of the transfection efficacy. Vector pUC19 was used as an empty donor plasmid for control. Plasmid maps (not drawn to scale) are illustrated. The donor plasmids were pIE100/1 (here referred to as pWT), specifying the authentic IE1 protein; pMut, specifying the mutated protein IE1-Y165C; and pRev, specifying the rescued protein IE1-C165Y. MIEPE, MIE promoter and enhancer; Ex, exon. The arrows indicate the direction of transcription; the arrowheads mark the location of the mutation in exon 4. Correct expression of the IE1 protein was inspected for all donor plasmids by IE1-specific immunofluorescence (not depicted). Luminescence data are expressed as RLU and are normalized for transfection efficacy by forming the quotient of firefly luciferase activity (RLUFL) and Renilla luciferase activity (RLURL). The solid circles and gray-shaded bars represent data from triplicate transfection cultures and their median value, respectively, for each experiment, with a total of three independent experiments performed for each type of donor plasmid tested. P values (two-sided), determined by the distribution-free (nonparametric) Wilcoxon-Mann-Whitney rank sum test (n1 = n2 = 9), are indicated.