Figure 3. ProFunc results for proteins of known function.
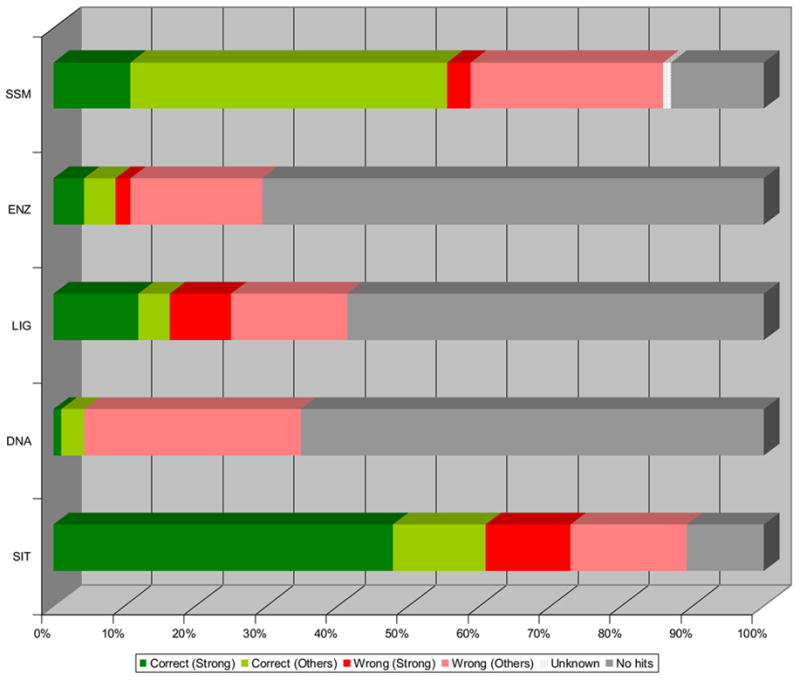
The 92 proteins classed as having “known function” in the MCSG dataset were analysed using ProFunc. The top hit (after parsing for release dates) was classified by success and strength of hit. Those hits to hypothetical proteins or members of families/domains of unknown function are classified as “unknown”. The structure-based methods used by ProFunc are as follows:
SSM – Secondary Structure Matching (MSDfold): fold comparison service.
ENZ – Enzyme template search (Catalytic Site Atlas data)
LIG – Ligand binding template search (Automatically generated templates)
DNA – DNA binding template search (Automatically generated templates)
SIT – SiteSeer (“Reverse template” method)
