Abstract
Reverse gyrase is a topoisomerase that introduces positive supercoils into DNA in an ATP-dependent manner. It is unique to hyperthermophilic archaea and eubacteria, and has been proposed to protect their DNA from damage at high temperatures. Cooperation between its N-terminal helicase-like and the C-terminal topoisomerase domain is required for positive supercoiling, but the precise role of the helicase-like domain is currently unknown. Here, the characterization of the isolated helicase-like domain from Thermotoga maritima reverse gyrase is presented. We show that the helicase-like domain contains all determinants for nucleotide binding and ATP hydrolysis. Its intrinsic ATP hydrolysis is significantly stimulated by ssDNA, dsDNA and plasmid DNA. During the nucleotide cycle, the helicase-like domain switches between high- and low-affinity states for dsDNA, while its affinity for ssDNA in the ATP and ADP states is similar. In the context of reverse gyrase, the differences in DNA affinities of the nucleotide states are smaller, and the DNA-stimulated ATPase activity is strongly reduced. This inhibitory effect of the topoisomerase domain decelerates the progression of reverse gyrase through the nucleotide cycle, possibly providing optimal coordination of ATP hydrolysis with the complex reaction of DNA supercoiling.
INTRODUCTION
Reverse gyrase is the only topoisomerase that introduces positive supercoils into DNA at the expense of ATP hydrolysis (1). It is unique to hyperthermophilic archaea and eubacteria. Deletion of the reverse gyrase gene in a hyperthermophilic archaeon leads to growth retardation at higher temperatures, but the organism is still able to survive at 90°C, demonstrating that reverse gyrase is not strictly required for hyperthermophilic life (2,3). The in vivo function of reverse gyrase is not clear. A heat-protective DNA chaperone activity (4) and a DNA renaturase activity (5) have been reported, indicating that reverse gyrase protects DNA from damage at high temperatures.
Reverse gyrase consists of an N-terminal helicase-like domain, fused to a C-terminal topoisomerase I domain. While most reverse gyrases are monomeric, a covalent connection between the two domains is not required for positive supercoiling activity. The Methanopyrus kandleri reverse gyrase is a dimeric enzyme, with the helicase-like and part of the topoisomerase domain in one subunit, and the remainder of the topoisomerase domain provided by the second subunit (6,7). Furthermore, an active reverse gyrase can be reconstituted by mixing separately produced helicase-like and topoisomerase domains (8; Hilbert,M. and Klostermeier,D., unpublished data). Nevertheless, a functional cooperation of helicase-like and topoisomerase domains is required for positive supercoiling by reverse gyrase (8,9).
The helicase-like domain shares the three-dimensional structure with helicases of the superfamily (SF) 2, namely two tandem RecA-folds (H1, H2) connected by a linker (Figure 1). In reverse gyrase, the subdomain H2 is interrupted by the so-called latch domain (H3) that shows homology to the region of the transcription termination factor Rho that is involved in RNA binding (10). The reverse gyrase helicase-like domain comprises all signature motifs of SF2 helicases, but the sequences of these motifs show large deviations from the consensus (Supplementary Figure S1). These sequence variations may be responsible for the lack of unwinding activity by reverse gyrase or the isolated helicase-like domain (8). Reverse gyrase does not catalyze relaxation in the absence of nucleotides, whereas the topoisomerase domain on its own can relax DNA in the absence of nucleotides, but lacks supercoiling activity (8,11). It has been suggested that the latch domain suppresses relaxation in reverse gyrase in the absence of nucleotides (12).
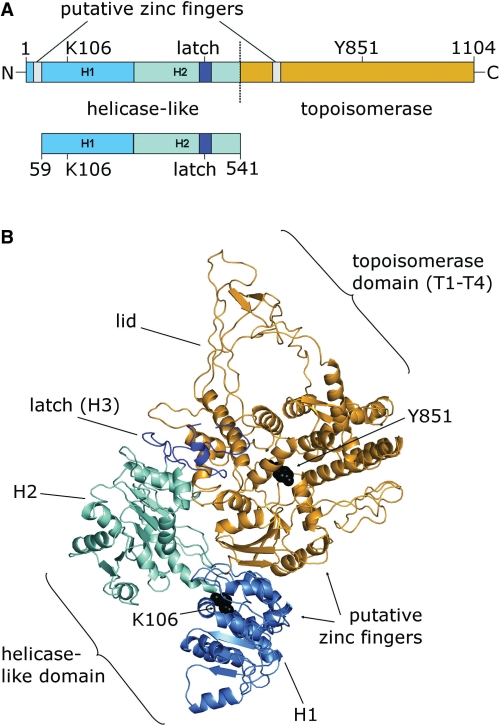
Figure 1.
Reverse gyrase and its helicase-like domain. (A) Full-length reverse gyrase, consists of an N-terminal helicase-like domain [H1, light blue, H2, cyan, H3 (latch), blue] and a C-terminal topoisomerase domain (orange). The helicase-like domain construct comprises the helicase-like domain (H1 and H2), and the latch-domain H3 that is inserted into H2. The positions of K106 in the Walker A motif and of the catalytic tyrosine, Y851 (black), and the putative zinc fingers (gray) are indicated. (B) Structure of A. fulgidus reverse gyrase [PDB-ID 1GKU, Ref. (10)], color-coded as in (A). The regions carrying the putative zinc fingers are missing in the electron density.
Based on the crystal structure of the reverse gyrase from Archaeoglobus fulgidus (10), a mechanism for positive supercoiling has been postulated that is based on a conformational change of the helicase-like domain as the initiating step, leading to a closure of the cleft between the two RecA-like subdomains. This conformational change may then allow for release of the latch domain, and for swinging-up of the ‘lid’ region in the topoisomerase domain, which is required for strand passage (Figure 1B). We have recently shown that such a conformational change occurs in SF2 helicases in response to the cooperative binding of ATP and nucleic acid substrates (13). Furthermore, we have demonstrated that reverse gyrase can use ATP or ATPγS as the energy source for positive supercoiling, suggesting that both nucleotides can power the conformational cycle (14). This ATPγS-dependent activity has also been observed for the translation factor eIF4A, a member of the SF2 helicase family (15), which further indicates mechanistic similarities of the reverse gyrase helicase-like domain and SF2 helicases. Although the helicase-like domain is a crucial element for ATP-dependent positive supercoiling by reverse gyrase, its role is currently not well understood. We present here the characterization of nucleotide binding, ATP hydrolysis, DNA binding and DNA-stimulation of the ATPase activity for the isolated helicase-like domain of Thermotoma maritima reverse gyrase and compare it to the properties of reverse gyrase. The helicase-like domain confers nucleotide-dependent DNA binding to reverse gyrase. The isolated domain is an efficient DNA-stimulated ATPase, but the topoisomerase domain in reverse gyrase exerts a moderating effect onto the ATPase activity, slowing down the nucleotide cycle by a factor of 10. This intra-molecular inhibition suggests that the helicase-like domain is harnessed by reverse gyrase to provide efficient coupling of ATPase activity with the supercoiling reaction.
MATERIALS AND METHODS
Cloning, mutagenesis, protein production and purification
The region encoding the helicase-like domain of reverse gyrase (E59-R541, rGyr_hel) was PCR-amplified from the full-length gene and cloned into pET28a using NcoI and XhoI restriction sites. Site-directed mutagenesis was performed according to the Quikchange protocol (Stratagene, La Jolla, CA, USA).
rGyr_hel was produced at 37°C in Escherichia coli Rosetta (DE3) (Invitrogen, Paisley, U.K) in autoinducing medium (16), and cells were harvested after 24 h. All purification steps were performed at room temperature. Cells were disrupted in a Microfluidizer in 50 mM Tris/HCl, pH 7.5, 1 M NaCl, 10 mM MgCl2, 10 μM Zn(OAc)2, 2 mM BME and the crude extract was cleared by centrifugation. The NaCl concentration of the supernatant was adjusted to 0.2 M, and it was applied to a SP sepharose column equilibrated in 50 mM Tris/HCl, pH 7.5, 0.2 M NaCl, 10 mM MgCl2, 10 μM Zn(OAc)2, 2 mM BME. rGyr_hel was eluted in a linear gradient from 0.2–1 M NaCl, dialyzed against 50 mM Tris/HCl, pH 7.5, 0.2 M NaCl, 10 mM MgCl2, 10 μM Zn(OAc)2, 2 mM BME, and applied to a Q sepharose column equilibrated in the same buffer. rGyr_hel was collected in the flowthrough. Final purification was achieved via size-exclusion chromatography on a calibrated S200 column in 50 mM Tris/HCl, pH 7.5, 0.2 M NaCl, 10 mM MgCl2, 10 μM Zn(OAc)2, 2 mM BME. rGyr_hel elutes as a monomer from a calibrated size-exclusion chromatography column (apparent molecular weight: 49.8 kDa, calculated: 56.3 kDa). Protein concentration was determined photometrically using the calculated extinction coefficient at 280 nm of 56 309 M−1 cm−1. From 1 l of bacterial culture, 4 mg of rGyr_hel with >98% purity (as judged from SDS–PAGE with Coomassie staining) were obtained. The pure protein was concentrated, shock-frozen in liquid nitrogen, and stored at −80°C. Full-length reverse gyrase was purified as described (14).
Adenine nucleotides and RNA and DNA substrates
Adenine nucleotides were purchased from Pharma Waldhof (Mannheim, Germany) or Jena Bioscience (Jena, Germany), and checked for impurities by reverse-phase HPLC on a C18 column in 0.1 M sodium phosphate, pH 6.5. Oligonucleotides for ssDNA or dsDNA substrates were obtained from Purimex (Grebenstein, Germany). The sequence of the 60-base ssDNA was 5′-(fluorescein)-AAGCCAAGCT TCTAGAGTCA GCCCGTGATA TTCATTACTT CTTATCCTAG GATCCCCGTT-3′, and the dsDNA substrate was formed by annealing the complementary strand. These substrates contain a preferred cleavage site (17,18) for reverse gyrase; cleavage occurs between the two underlined nucleotides. PolyU-RNA was purchased from Sigma (Hamburg, Germany).
ATPase assays
Steady state ATPase activity was measured in a spectroscopic enzymatic assay that couples ADP production to the oxidation of NADH as described (14). Assay conditions were 50 mM Tris/HCl, pH 7.5, 0.15 M NaCl, 10 mM MgCl2, 100 μM Zn(OAc)2, 2 mM BME, 0.4 mM PEP and 0.2 mM NADH, 23 μg ml−1 LDH, 37 μg ml−1 PK. ATPase activity at 75°C was determined in 50 mM Tris/HCl, pH 7.5, 0.15 M NaCl, 10 mM MgCl2, 100 μM Zn(OAc)2, 2 mM BME, 10% (w/v) PEG 8000 by mixing 1 μM enzyme, 2 mM ATP and the respective nucleic acid substrate, taking aliquots at different time points, and analyzing the nucleotide composition by reverse-phase HPLC as described (14).
Fluorescence measurements
Dissociation constants of rGyr_hel/nucleotide complexes were determined in fluorescence equilibrium titrations at 37°C using 1 μM of the fluorescent ADP analog mantADP (19) and in competitive titrations of the mantADP/rGyr_hel complex with ADP, ATPγS, ADPNP, ADPCP and ATP, and analyzed as described (14).
Dissociation constants of reverse gyrase/DNA complexes were determined using 5′-fluorescein-labeled DNA and the steady-state anisotropy of fluorescein as a probe for binding. The DNA concentrations were 25 nM for titrations with rGyr_hel, and 10 nM for rGyr_fl, unless stated otherwise. Dissociation constants were obtained by analysis of the data using the solution of a quadratic equation describing a one-site binding model as described (14), or with the Hill equation.
Helicase assay
Helicase activity was tested via the displacement of a 10-mer from a 10/50-mer substrate (2 nM) by 10 μM rGyr_hel in 20 mM HEPES/NaOH, pH 7.5, 70 mM KCl, 1 mM MgOAc, 10% (v/v) glycerol, 2 mM DTT and 1 mg/ml BSA as described for eIF4A (20). The ATP concentration was 2 mM.
RESULTS
Nucleotide binding and ATPase properties of the helicase-like domain
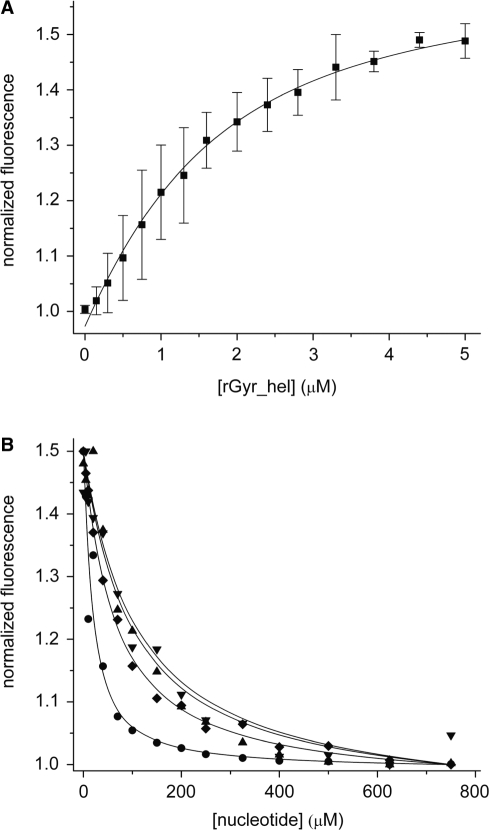
The role of the helicase-like domain for DNA supercoiling by reverse gyrase is currently unknown. A detailed understanding of nucleotide binding and hydrolysis by the helicase-like domain and its interaction with DNA, isolated and in the context of reverse gyrase, is a pre-requisite to delineate its function within reverse gyrase. To this end, we subcloned a DNA fragment coding for amino acids 59–541 of T. maritima reverse gyrase (rGyr_hel). This construct comprises the two RecA-like domains H1 and H2, and the so-called latch domain H3, but lacks the N-terminal putative zinc finger (Figure 1). The nucleotide binding properties of rGyr_hel were investigated in fluorescent equilibrium titrations using the fluorescent ADP analog mantADP (Figure 2), which we previously used as a probe for nucleotide binding to reverse gyrase (14). Upon binding to rGyr_hel, the mant fluorescence increases 1.5-fold (Figure 2A). The fluorescence signal returns to the value for free mantADP upon displacement with ADP, confirming that mantADP binds to the ADP binding site and is thus suitable to study nucleotide binding to rGyr_hel. The Kd value of the mantADP/rGyr_hel complex determined from the titration curves is 1.1 ± 0.1 μM. In the displacement titration with ADP (Figure 2B), a Kd value of 2.6 ± 0.8 μM was determined for the ADP complex. The Kd value of the rGyr_hel/AMP complex is three orders of magnitude higher (1600 ± 231 μM), demonstrating that interactions with the β-phosphate provide high affinity binding of adenine nucleotides to rGyr_hel. The ATP analogs ATPγS, ADPNP and ADPCP are bound less tightly than ADP (Figure 2B), with Kd values for the complexes of 10.9 ± 0.8 μM (ATPγS), 20 ± 4.9 μM (ADPNP) and 18 ± 3.8 μM (ADPCP), respectively, indicating that the binding energy from additional interactions with the γ-phosphate is converted into conformational changes. The corresponding Kd values for the full-length reverse gyrase (rGyr_fl) are virtually identical in the case of the mantADP, ADP and AMP complexes, and of the ATP analog ATPγS, and 2- to 4-fold higher for the complexes with the nonhydrolyzable ATP analogs ADPNP and ADPCP (14). The similar Kd values confirm that all determinants for nucleotide binding are contained in the helicase-like domain. A summary of the Kd values for rGyr_fl and rGyr_hel is given in Table 1.
Figure 2.
Interaction of rGyr_hel with nucleotides. (A) Titration of 1 μM mantADP with rGyr_hel. Description of the binding curve using a one-site binding model yields a Kd of the mantADP/rGyr_hel complex of 1.1 ± 0.1 μM. (B) Displacement titrations of the mantADP/rGyr_hel complex with ADP (circles), ADPCP (triangle), ADPNP (inverted triangle), and ATPγS (diamond). The Kd values are summarized in Table 1.
Table 1.
Dissociation constants for nucleotide complexes of full-length reverse gyrase and the helicase-like domain
| Nucleotide | rGyr_fl Kd (μM) | rGyr_hel Kd (μM) | rGyr_fl K106Q/Y851F Kd (μM) | rGyr_hel K106Q Kd (μM) |
|---|---|---|---|---|
| mantADP | 1.0 ± 0.1a | 1.1 ± 0.1 | 22 ± 2 | 67 ± 5 |
| AMP | 1549 ± 375 | 1600 ± 231 | ND | ND |
| ADP | 1.6 ± 0.2a | 2.6 ± 0.8 | 36 ± 5 | 150 ± 85 |
| ATP | ND | ND | 98 ± 12 | 570 ± 300 |
| ATPγS | 14.3 ± 1.5a | 10.9 ± 0.8 | ND | ND |
| ADPNP | 43.2 ± 2.0a | 19.8 ± 4.9 | ND | ND |
| ADPCP | 74.7 ± 7.3 | 17.5 ± 3.8 | ND | ND |
aData from Ref. (14).
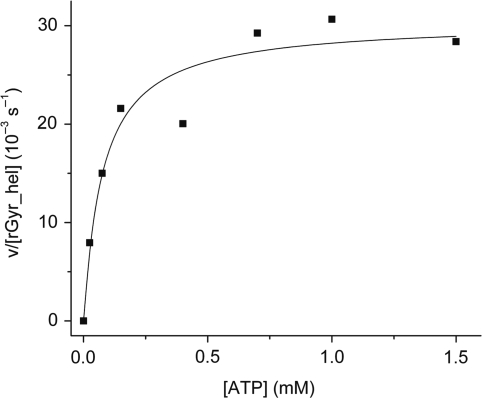
In a steady state ATPase assay, rGyr_hel exhibits a low intrinsic ATPase activity (Figure 3). As with rGyr_fl (14), the rate constant kcat of ATP hydrolysis by rGyr_hel is independent of the protein concentration (<10 μM, data not shown), consistent with a monomer as the active species. The dependence of the ATP hydrolysis rate on ATP concentration follows Michaelis–Menten behavior, with a kcat for ATP hydrolysis of 30 (± 2) × 10−3 s−1, and a KM value for ATP of 77 ± 23 μM. The corresponding values for reverse gyrase are very similar with a kcat of 20 (± 0.8) × 10−3 s−1 and a KM of 44 ± 6 μM (14).
Figure 3.
Steady state ATPase activity of rGyr_hel. rGyr_hel is a Michaelis–Menten enzyme with a kcat of 30 (±2) × 10−3 s−1, and a KM,ATP of 77 ± 23 μM.
These results demonstrate that the nucleotide binding properties and the intrinsic ATPase activity of reverse gyrase are a property of the helicase-like domain and are not significantly affected by the topoisomerase domain.
Stimulation of the rGyr_hel and rGyr_fl ATPase activities by nucleic acids
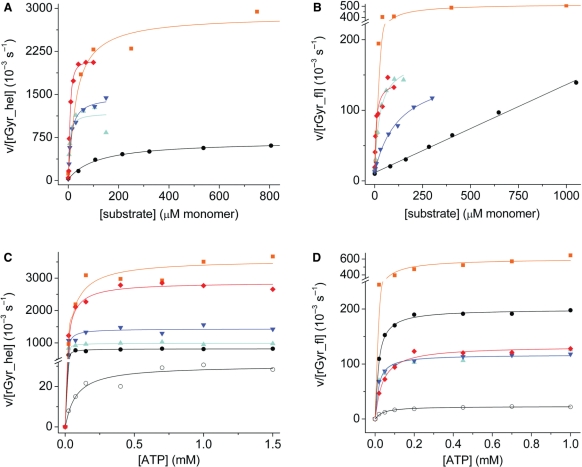
Reverse gyrases are DNA-stimulated ATPases (21). To investigate the effect of nucleic acid substrates on the ATPase activity of rGyr_hel, steady state ATPase assays were performed in the presence of a 60-base ssDNA substrate, a 60 bp dsDNA, pUC18 plasmid DNA, and polyU-RNA at saturating ATP concentrations (Figure 4A). All substrates led to a significant acceleration of ATP hydrolysis. Saturating concentrations of ssDNA or linear dsDNA increased the kcat by a factor of 40–50, with a slightly higher stimulation by dsDNA. The apparent KM values are 0.07 ± 0.04 μM for ssDNA (corresponding to 4 μM in terms of bases) and 0.18 ± 0.03 μM for dsDNA (11 μM base pairs), respectively. In the presence of negatively supercoiled pUC18 plasmid, the kcat value increased 22-fold (KM 0.046 ± 0.006 μM, corresponding to 124 μM base pairs).
Figure 4.
Effect of nucleic acid substrates on the steady state ATPase activity of rGyr_hel and rGyr_fl. (A and B) The effect of nucleic acid substrates on the ATPase rate at saturating (1 mM) ATP. (C and D) The ATP-dependence of the ATPase rate at saturating concentrations of nucleic acids. Substrates are indicated as follows: ssDNA (cyan, triangles), dsDNA (blue, inverted triangles), pUC18 (black, circles), polyU-RNA (orange, squares) and heparin (red, diamond). The open symbols in (C and D) indicate data in the absence of DNA. All steady state ATPase parameters are summarized in Tables 2 and 3. (A) Stimulation of the rGyr_hel ATPase by nucleic acids and heparin. The ATPase activity is significantly stimulated by all substrates. (B) Stimulation of the rGyr_fl ATPase by nucleic acids and heparin. The ATPase activity is significantly stimulated by all substrates, but in contrast to rGyr_hel, no saturation is observed with pUC18. (C) Cooperativity between ATP and nucleic acid binding in rGyr_hel. The steady state ATPase activity was measured as a function of ATP concentration in the presence of saturating concentrations of ssDNA (2 μM,), dsDNA (2.5 μM), pUC18 (300 nM), polyU-RNA (1.5 mM monomers) and heparin (40 μM monomers). (D) Cooperativity between ATP and nucleic acid binding in rGyr_fl. The steady state ATPase activity was measured as a function of ATP concentration in the presence of saturating concentrations of ssDNA (2 μM,), dsDNA (5 μM), pUC18 (300 nM), polyU-RNA (0.5 mM monomers) and heparin (60 μM monomers).
Interestingly, polyU-RNA also stimulated the intrinsic ATPase activity of rGyr_hel tremendously (∼100-fold). With 25 μM (bases), the apparent KM value for polyU-RNA was in the same range as for the pUC18 plasmid. PolyU-RNA binding might reflect nonspecific interactions with the negatively charged phosphoribose backbone, and we therefore repeated the steady state ATPase assay in the presence of increasing concentrations of heparin as a model for a negatively charged polymer. Indeed, heparin increased the kcat of rGyr_hel for ATP hydrolysis ∼80-fold and thus had a comparable effect to the polyU-RNA. The KM value for heparin was 0.20 ± 0.05 μM (8 μM monomeric units) and thus also similar to polyU.
To compare ATPase properties of the helicase-like domain with reverse gyrase, the steady state ATPase rates in the presence of ssDNA, dsDNA, pUC18 and polyU-RNA were also determined for rGyr_fl (Figure 4B). As with rGyr_hel, linear ssDNA and dsDNA showed a similar effect on the ATPase rate of rGyr_fl. Importantly, the stimulation of the ATPase activity by ssDNA and dsDNA was only 7- to 8-fold. The ATPase rate of rGyr_fl increases linearly up to a pUC18 concentration of 400 nM (1.1 mM bases), and saturation is not observed (14). In contrast to rGyr_hel, pUC18 only showed a moderate stimulation of the rGyr_fl ATPase rate (7-fold at 400 nM pUC18, compared to 22-fold at saturation for rGyr_hel). As with rGyr_hel, polyU-RNA showed the highest degree of stimulation of the reverse gyrase ATPase activity, with kcat increased 25-fold (KM 30 μM bases), and heparin showed a 6-fold stimulation (KM 6 μM monomeric units). These data indicate that interactions with nonspecific substrates are similar for rGyr_hel and rGyr_fl. The kcat values for rGyr_fl and rGyr_hel in the absence and presence of nucleic acids and the apparent KM values are summarized in Table 2.
Table 2.
Steady state ATPase parameters for rGyr_hel and rGyr_fl, and the influence of nucleic acid substrates
| Nucleic acid | kcat (10−3 s−1) | KM,DNA (μM) | KM (μM base/bp) | |
|---|---|---|---|---|
| rGyr_hel | – | 30 ± 2 | NA | NA |
| ssDNA | 1160 ± 188 | 0.07 ± 0.04 | 4.2 ± 2.4 | |
| dsDNA | 1450 ± 64 | 0.18 ± 0.03 | 10.8 ± 1.8 | |
| pUC18 | 673 ± 21 | 0.046 ± 0.006 | 124 ± 16 | |
| polyU-RNA | 3264 ± 304 | NA | 25 ± 10 | |
| Heparin | 2420 ± 160 | 0.20 ± 0.05 | 8.0 ± 0.04 | |
| rGyr_fl | – | 20 ± 0.8 | NA | NA |
| ssDNA | 160 ± 6.7 | 0.45 ± 0.06 | 27 ± 4 | |
| dsDNA | 148 ± 9.1 | 2.2 ± 0.30 | 129 ± 18 | |
| pUC18 | No saturation for conc. <400 nM | |||
| polyU-RNA | 507 ± 27 | NA | 30 ± 7 | |
| Heparin | 120 ± 10 | 0.15 ± 0.06 | 6 ± 2.4 | |
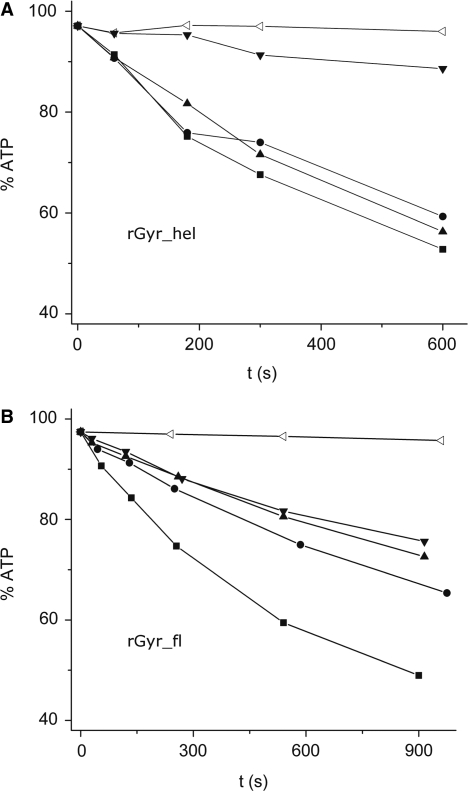
Reverse gyrase is an enzyme unique to hyperthermophilic organisms, which thrive at temperatures of 75°C and higher. To investigate if the results from experiments at 37°C reflect the properties of reverse gyrase at its optimum temperature, the ATPase activity was determined at 75°C in the absence of DNA, and in the presence of ssDNA, dsDNA and pUC18 (Figure 5). In the absence of DNA, the ATPase activities of rGyr_fl and rGyr_hel are similar, with initial rates of 0.28 μM s−1 (rGyr_hel) and 0.48 μM s−1 (rGyr_fl). While we have determined the affinities for DNA substrates at 37°C and could therefore ensure saturating DNA concentrations in ATPase assays, a quantitative comparison of the ATPase stimulation at 75°C is difficult as the corresponding Kd values cannot easily be determined, and the DNA concentrations may not be saturating. In addition, the thermodynamic stability of the 60 bp duplex is limited (Tm = 70°C), and at 75°C more than 50% single strand are expected. Qualitatively, the rGyr_hel ATPase activity at 75°C is stimulated to a similar extent by ssDNA and dsDNA at 75°C, and about 2-fold less by pUC18. Overall, the ATPase activity is increased only 3-fold by DNA substrates, compared to 50-fold at 37°C. The rGyr_fl ATPase is most efficiently stimulated by ssDNA, and to a smaller extent by dsDNA or pUC18. As observed at 37°C, the overall stimulation is smaller for rGyr_fl than for rGyr_hel. Thus, the rGyr_hel and rGyr_fl ATPase activities at 75°C show the same response to DNA as at 37°C.
Figure 5.
DNA-stimulated ATPase activity at 75°C. ATPase activity of rGyr_hel (A) and rGyr_fl (B) at 75°C in the absence of enzyme (open triangles), in the presence of 1 μM enzyme without DNA (inverted triangles) and in the presence of ssDNA (100 nM, squares), dsDNA (100 nM, circles), or pUC18 (15 nM, triangles). The ATP concentration was 1 mM.
In summary, the basal ATP hydrolysis rates are very similar for rGyr_hel and rGyr_fl, confirming that all determinants for ATP binding and hydrolysis are confined to the helicase-like domain. The intrinsic ATPase of rGyr_hel is efficiently stimulated by ssDNA, dsDNA and plasmid DNA. All substrates also stimulate the intrinsic ATP hydrolysis of rGyr_fl, but generally to a lesser extent. These data point towards an inhibitory effect of the topoisomerase domain on the ATPase activity of the helicase-like domain in the presence of DNA substrates. The apparent KM values for DNA are higher for rGyr_fl than for rGyr_hel. This effect seems to be slightly more pronounced for dsDNA, leading to a 5-fold higher apparent KM for dsDNA over ssDNA for rGyr_fl, compared to an only 2.5-fold difference for rGyr_hel. Both enzymes thus appear to interact more tightly with ssDNA. Interestingly, the apparent KM values for polyU-RNA and heparin are identical for rGyr_fl and rGyr_hel, indicating similar interactions with nonspecific substrates.
Coupling between DNA and ATP binding to rGyr_hel and rGyr_fl
The interaction of SF2 helicases with ATP and their DNA substrate is cooperative: ATP binding increases the affinity for the DNA substrate, and vice versa (13,22–24). To test whether any of the nucleic acid substrates shows such a cooperative effect on ATP binding to rGyr_hel or rGyr_fl, we determined the effect of DNA on KM values for ATP by measuring the steady state ATPase rate with increasing concentrations of ATP and saturating concentrations of the nucleic acid (Figure 4C and D). In all cases, the kcat values are in good agreement with the values in the DNA-dependent experiments at saturating ATP concentrations (Table 2), confirming that saturation is reached in both series of experiments. In the presence of ssDNA, the apparent KM,ATP value of rGyr_hel is decreased 16-fold, demonstrating cooperative binding of ATP and ssDNA to rGyr_hel. For dsDNA, this cooperativity is lower than for ssDNA, with KM,ATP decreased 7-fold. The effect of pUC18 plasmid is intermediate, with a 10-fold decrease in the apparent KM,ATP. Hence, all DNA substrates promote ATP binding to rGyr_hel. PolyU-RNA and heparin only affect the apparent KM,ATP slightly (1.8-fold and 2.5-fold, respectively) and thus do not show significant cooperativity. These data corroborate the notion that the stimulating effect of polyU-RNA and heparin on the ATPase activity of rGyr_hel is due to a nonspecific interaction.
For rGyr_fl, limited cooperativity of DNA and ATP binding was detected (Figure 4D). The reduction of the KM,ATP was 3.8-fold for ssDNA and 2.8-fold for dsDNA. Hence, as for rGyr_hel, the cooperativity with ATP binding is highest for ssDNA. However, the cooperativity of DNA and ATP binding to rGyr_fl is smaller for ssDNA and dsDNA compared to rGyr_hel. For pUC18, it was not possible to perform the experiment under saturating conditions. At 300 nM pUC18, KM,ATP was reduced 2.8-fold. This is similar to the effect of linear dsDNA, whereas for rGyr_hel, pUC18 shows intermediate cooperativity to ssDNA and dsDNA. However, a more pronounced effect of pUC18 on KM,ATP for rGyr_fl at higher plasmid concentrations cannot be excluded. Overall, rGyr_fl shows cooperative interactions with DNA and ATP, but to a lesser extent than rGyr_hel. PolyU-RNA and heparin had a negligible effect on ATP binding (1.4-fold and 1.2-fold, respectively), consistent with a nonspecific interaction. The kcat and KM,ATP values are summarized in Table 3.
Table 3.
Steady state ATPase parameters for rGyr_hel and rGyr_fl: the influence of nucleic acid substrates on KM,ATP
| Nucleic acid (saturating) | kcat (10−3 s−1) | KM,ATP (μM) | |
|---|---|---|---|
| rGyr_hel | – | 30 ± 2 | 77 ± 23 |
| ssDNA | 992 ± 12 | 4.8 ± 1.0 | |
| dsDNA | 1435 ± 57 | 11 ± 4 | |
| pUC18 | 818 ± 10 | 7.4 ± 1.1 | |
| polyU-RNA | 3402 ± 79 | 42 ± 5.5 | |
| Heparin | 2869 ± 63 | 31 ± 4 | |
| rGyr_fl | – | 20 ± 0.8 | 44 ± 6 |
| ssDNA | 108 ± 2.1 | 12 ± 1.5 | |
| dsDNA | 117 ± 1.6 | 16 ± 1.5 | |
| pUC18 (300 nM) | 199 ± 1.6 | 15.6 ± 0.9 | |
| polyU-RNA | 565 ± 19 | 32 ± 5.1 | |
| Heparin | 133 ± 7 | 37 ± 6.7 |
Altogether, the interaction of rGyr_hel and rGyr_fl with polyU-RNA or heparin and ATP is noncooperative and most likely reflects a nonspecific interaction. Substantial cooperativity is observed for ssDNA, dsDNA or plasmid DNA and ATP binding to rGyr_hel. rGyr_fl also binds ssDNA or dsDNA and ATP cooperatively, but the effect of the DNA on KM,ATP is only 3- to 4-fold, compared to 7- to 16-fold for rGyr_hel. Again, this points towards a moderating effect of the topoisomerase domain on the helicase-like domain in full-length reverse gyrase.
Influence of the nucleotide state on DNA binding to rGyr_hel
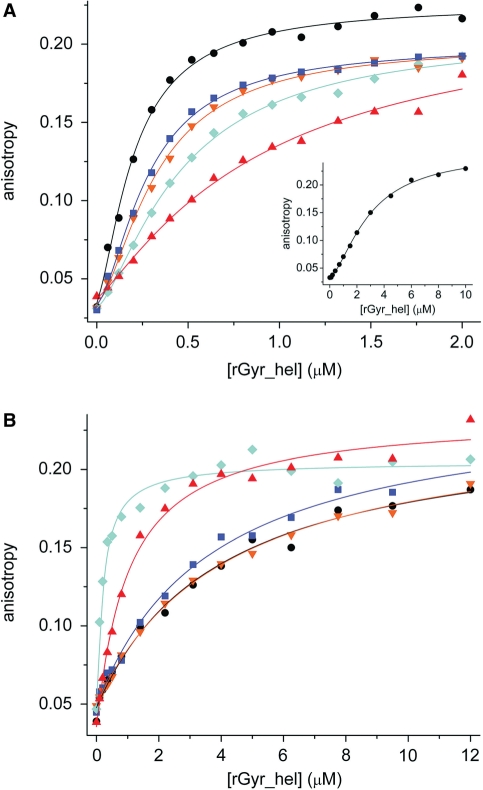
During their nucleotide cycle, SF2 helicases switch between low affinity and high affinity states for their nucleic acid substrate (22,25,26). We therefore investigated the influence of the nucleotide state of rGyr_hel and rGyr_fl on their interaction with DNA. To this end, the affinity of rGyr_hel for nucleic acids was determined in fluorescence anisotropy titrations of a 5′-fluorescein-labeled 60-base ssDNA or a fluorescein-labeled 60 bp dsDNA in the absence of nucleotides, and in the presence of saturating concentrations of ADP, ADPCP, ADPNP and ATPγS (Figure 6, Table 4).
Figure 6.
DNA binding and coupling to nucleotide binding (rGyr_hel). Titrations of DNA with rGyr_hel in the absence of nucleotide (black, circles), and in the presence of ADP (100 μM, blue, squares), ADPCP (500 μM, orange, inverted triangles), ATPγS (500 μM, red, triangles) and ADPNP (500 μM, cyan, diamonds). All Kd values and Hill coefficients are summarized in Table 4. (A) ssDNA (25 nM). The binding curves are described with the Hill equation. The inset shows the stoichiometric titration of 2 μM ssDNA with rGyr_hel. (B) dsDNA (25 nM). The binding curves are described with a one-site binding model.
Table 4.
DNA binding properties of different nucleotide states of rGyr_hel
| Kd (μM) | nucleotide-free | ADP state | ATP state | ||
|---|---|---|---|---|---|
| ssDNA | |||||
| wt | 0.20 ± 0.03a | ADP | 0.31 ± 0.04a | ADPNP | 0.60 ± 0.07a |
| 0.20 ± 0.01b | 0.28 ± 0.01b | 0.46 ± 0.03b | |||
| n = 1.4 ± 0.1 | n=1.6 ± 0.1 | n=1.4 ± 0.1 | |||
| ADPCP | 0.38 ± 0.05a | ATPγS | 1.2 ± 0.2a | ||
| 0.33 ± 0.01b | 0.92 ± 0.21b | ||||
| n=1.6 ± 0.1 | n=1.2 ± 0.2 | ||||
| K106Q | 0.18 ± 0.03a | 0.25 ± 0.03a | 0.19 ± 0.02a | ||
| 0.18 ± 0.03b | 0.24 ± 0.02b | 0.20 ± 0.02b | |||
| n = 1.3 ± 0.2 | n=1.3 ± 0.1 | n=1.1 ± 0.1 | |||
| dsDNA | |||||
| wt | 3.9 ± 0.6 a | ADP | 3.7 ± 0.5a | ADPNP | 0.19 ± 0.03a |
| ADPCP | 4.2 ± 0.3a | ATPγS | 1.0 ± 0.1 a | ||
| K106Q | 3.8 ± 0.6 a | 2.9 ± 0.6 a | 1.7 ± 0.2 a | ||
aOne-site.
bHill analysis.
First, we characterized binding of ssDNA to rGyr_hel (Figure 6A). The Kd value for the rGyr_hel/ssDNA complex was 0.20 ± 0.03 μM in the absence of nucleotide, and varied only moderately with the nucleotide state. The nonhydrolyzable ATP analogs ADPNP and ATPγS showed the largest effect on ssDNA binding, with 3-fold (ADPNP) or 6-fold (ATPγS) increased Kd values, respectively, indicating that the affinity for ssDNA is slightly reduced in the ATP state of rGyr_hel. The ADP state of rGyr_hel binds ssDNA 1.5-fold less tightly than the nucleotide-free state, but 2- to 4-fold more tightly than the ATP-state (mimicked by ADPNP and ATPγS). ADPCP has a similar effect on ssDNA binding as ADP, suggesting that ADPCP is not a suitable mimic for ATP.
When the concentration of ssDNA in the titration with rGyr_hel was increased to 2 µM, a sigmoidal binding curve was obtained, indicating that more than one rGyr_hel molecule interacts with each ssDNA (Figure 6A, inset). The data were well-described by the Hill equation, with a Hill coefficient of 1.6 ± 0.1, and a Kd of 2.9 ± 0.1 μM. Analysis of all ssDNA titration curves with the Hill equation gave similar Kd values as the analysis with the one-site binding model (Table 4), and Hill coefficients of 1.2–1.6, indicating that the cooperativity is moderate.
Next, binding of rGyr_hel to dsDNA was characterized (Figure 6B, Table 4). DsDNA was bound 10- to 20-fold less tightly by rGyr_hel than ssDNA, consistent with the observed higher KM value for dsDNA. The Kd value for dsDNA was ∼3.9 μM in the absence of nucleotide, or in the presence of ADP or ADPCP. Only in the presence of ADPNP or ATPγS was a significant change observed: the Kd value of the dsDNA/rGyr_hel complex was decreased 20-fold by ADPNP, or 4-fold by ATPγS. This decrease is consistent with ADPNP and ATPγS mimicking the ATP state. Overall, the ATP state of rGyr_hel thus binds dsDNA 4- to 20-fold more tightly than the ADP state.
Comparison of DNA binding to different nucleotide states (Table 4) reveals that the nucleotide-free form of rGyr_hel shows 20-fold higher affinity for ssDNA over dsDNA. In the ATP state, the affinity for ssDNA remains similar, but the dsDNA affinity increases compared to the nucleotide-free state. Thus, the ATP state exhibits similar affinities for ssDNA and dsDNA. In the ADP state, the affinity for ssDNA is similar to the nucleotide-free or ATP states, but the affinity for dsDNA is reduced compared to the ATP state. As a consequence, ssDNA binding is favored 11- to 13-fold in the ADP state. Hence, during the nucleotide cycle, rGyr_hel does not discriminate between ssDNA and dsDNA prior to ATP hydrolysis, but will preferentially interact with ssDNA after hydrolysis.
Due to thermodynamic coupling, the observed increased affinity of the ATP state for dsDNA requires a reciprocal effect of DNA on ATP binding. Comparison with the steady state ATPase data (Figure 4A and C, Table 3) shows that this coupling is indeed present: the KM,ATP of rGyr_hel is decreased 7-fold in the presence of dsDNA, which is in good agreement with the 4- to 20-fold increased dsDNA affinity in the ATP state of rGyr_hel. The comparison for the ssDNA seems to be inconsistent: the ATP state of rGyr_hel binds ssDNA 3- to 6-fold less tightly than the nucleotide-free form, but KM,ATP is decreased 16-fold when ssDNA is bound. However, the observed differences most likely reflect differences of the ADPNP or ATPγS states compared to the ATP state. This is partly supported by experiments with a hydrolysis-deficient mutant (see below), where ATP binding does not reduce the ssDNA affinity.
Influence of the nucleotide state on DNA binding to rGyr_fl
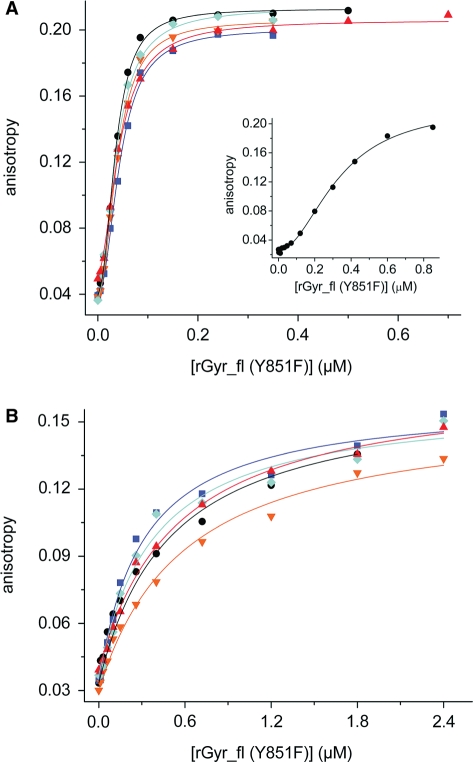
To test whether the helicase-like domain also acts as a nucleotide-dependent switch with different affinities for dsDNA in the ATP and ADP states in reverse gyrase, we performed DNA binding experiments with rGyr_fl (Figure 7). Quantification of DNA binding to rGyr_fl is complicated by the existence of several potential DNA binding sites that could be located in the helicase-like domain, in the cleft of the topoisomerase domain near the catalytic tyrosine, and possibly at the two putative zinc fingers. To ensure equilibrium conditions, we used the Y851F mutant of reverse gyrase in DNA binding experiments, which is deficient in covalent binding to the DNA.
Figure 7.
DNA binding and coupling to nucleotide binding (rGyr_fl). Titration of DNA with rGyr_fl in the absence of nucleotide (black, circles), and in the presence of ADP (100 μM, blue, squares), ADPCP (500 μM, orange, inverted triangles), ATPγS (500 μM, red, triangles) and ADPNP (500 μM, cyan, diamonds). All Kd values and Hill coefficients are summarized in Table 5. (A) ssDNA (10 nM). The binding curves are described with the Hill equation. The inset shows the stoichiometric titration of 300 nM ssDNA with rGyr_fl. (B) dsDNA (10 nM). The binding curves are described with a one-site binding model.
Again, we first addressed binding of ssDNA to rGyr_fl (Figure 7, Table 5). In a titration of the 60-base ssDNA with rGyr_fl(Y851F), a sigmoidal dependence of the fluorescence anisotropy on the concentration of enzyme was observed (Figure 7A). The binding isotherm was well-described by the Hill equation, with a Hill coefficient of 2.3 (± 0.2) and a Kd of 36 ± 1 nM for nucleotide-free rGyr_fl(Y851F), pointing towards cooperative binding of at least two reverse gyrase molecules to the DNA. To confirm the existence of cooperativity, the titration was repeated at 300 nM of ssDNA, where the sigmoidality became much more prominent (Figure 7A, inset). Description with the Hill equation yielded Kd = 0.33 ± 0.02 μM, and n = 2.0 ± 0.14). In the presence of saturating concentrations of ADP, ADPCP, ADPNP or ATPγS, binding isotherms for ssDNA to rGyr_fl remained sigmoidal (Figure 7A), with Hill coefficients from 1.8 ± 0.10 to 2.1 ± 0.16, again consistent with two reverse gyrase molecules binding to one ssDNA molecule.
Table 5.
DNA binding properties of different nucleotide states of rGyr_fl
| Kd (μM) | nucleotide-free | ADP state | ATP state | ||
|---|---|---|---|---|---|
| ssDNA | |||||
| Y851F | 0.035 ± 0.005a | ADP | 0.060 ± 0.015a | ADPNP | 0.041 ± 0.009a |
| 0.036 ± 0.001b | 0.045 ± 0.002b | 0.038± 0.002b | |||
| n = 2.3 ± 0.2 | n = 2.1 ± 0.2 | n = 2.1 ± 0.2 | |||
| ADPCP | 0.049 ± 0.012a | ATPγS | 0.046 ± 0.008 | ||
| 0.039 ± 0.001b | 0.041 ± 0.001b | ||||
| n = 2.1 ± 0.1 | n = 1.8 ± 0.1 | ||||
| K106Q/Y851F | 0.047 ± 0.005a | 0.053 ± 0.007a | 0.037 ± 0.004a | ||
| 0.044 ± 0.003b | 0.044 ± 0.002b | 0.035 ± 0.002b | |||
| n = 1.4 ± 0.1 | n = 1.5 ± 0.1 | n = 1.4 ± 0.1 | |||
| dsDNA | |||||
| Y851F | 0.49 ± 0.06a | ADP | 0.25 ± 0.04a | ADPNP | 0.33 ± 0.07a |
| ADPCP | 0.59 ± 0.06a | ATPγS | 0.53 ± 0.07a | ||
| K106Q/Y851F | 0.71 ± 0.11a | 0.46 ± 0.07a | 0.49 ± 0.09a | ||
aone-site.
bHill analysis.
Overall, rGyr_fl binds ssDNA 6-fold more tightly than rGyr_hel. In the presence of ADP, the interaction with ssDNA is only slightly affected, and it is virtually unchanged in the presence of nonhydrolyzable ATP analogs. Consequently, the affinity of rGyr_fl for ssDNA is not significantly modulated by the nucleotide state.
Next, the interaction of rGyr_fl(Y851F) with dsDNA was characterized (Figure 7, Table 5). Interestingly, the cooperativity was less pronounced for rGyr_fl binding to dsDNA (Figure 7B). In this case, the binding isotherm could be described by a simple one-site model with a Kd of 490 ± 60 nM. Thus, rGyr_fl interacts 8-fold more tightly with dsDNA than rGyr_hel. dsDNA bound to rGyr_fl was displaced upon addition of an excess of ssDNA (data not shown), demonstrating that both types of nucleic acids interact with the same binding site.
In the presence of nucleotides, the cooperativity of dsDNA binding appeared to increase slightly, as indicated by a slightly improved fit with the Hill equation. Hill coefficients varied between 0.7 and 1.7, consistent with a lower cooperativity compared to ssDNA (or a reduced number of binding sites). However, the experimental data were already reasonably well described with the one-site binding model. The presence of ADP, ADPNP, ADPCP or ATPγS affects the affinity of rGyr_fl for dsDNA <2-fold.
Comparison of DNA binding to different nucleotide states of rGyr_fl (Table 5) shows that the nucleotide-free form binds dsDNA 14-fold less tightly than ssDNA. In the ATP or state, ssDNA or dsDNA binding are not affected. However, the ADP state binds dsDNA 2-fold more tightly than the nucleotide-free and the ATP states. In the nucleotide cycle, both the ATP and the ADP state bind ssDNA more tightly than dsDNA. The preference for ssDNA is slightly reduced after hydrolysis (from 10-fold in the ATP state to 4-fold in the ADP state). The small effect of nucleotides on DNA binding to rGyr_fl is consistent with the small effect of ssDNA and dsDNA on ATP binding according to the steady state ATPase data (Figure 4B and D, Table 3).
In summary, the ATP state of rGyr_hel binds ssDNA and dsDNA with equal affinities, whereas the ADP state interacts preferentially with ssDNA. rGyr_fl binds DNA ∼10-fold more tightly than rGyr_hel, but the differences between the nucleotide states are far less pronounced. Both the ATP and the ADP states bind preferentially to ssDNA.
DNA binding to different nucleotide states of a hydrolysis-deficient mutant of rGyr_fl and rGyr_hel
As complexes with the nonhydrolyzable ATP analogs might not perfectly mimic the ATP states, we analyzed the effect of nucleotides on the interaction with DNA using hydrolysis-deficient mutants of rGyr_hel and rGyr_fl. A conserved lysine in the Walker A motif of ATPases contacts the β- and γ-phosphates of ATP, and stabilizes negative charges during ATP hydrolysis. In the rGyr_fl(K106Q) mutant, ATP hydrolysis is abolished, but nucleotides are still bound, albeit with reduced affinity (14). The rGyr_fl(K106Q/Y851F) double mutant was created to stably generate ATP and ADP states of reverse gyrase that do not covalently bind to DNA. The absence of ATP hydrolysis was confirmed, and the Kd values of the nucleotide complexes were determined to be 22 ± 2 μM (mantADP), 36 ± 5 μM (ADP) and 98 ± 12 μM (ATP) in equilibrium titrations (Table 1). Overall, nucleotide binding to rGyr_fl(K106Q/Y851F) is reduced ∼20-fold compared to wild-type.
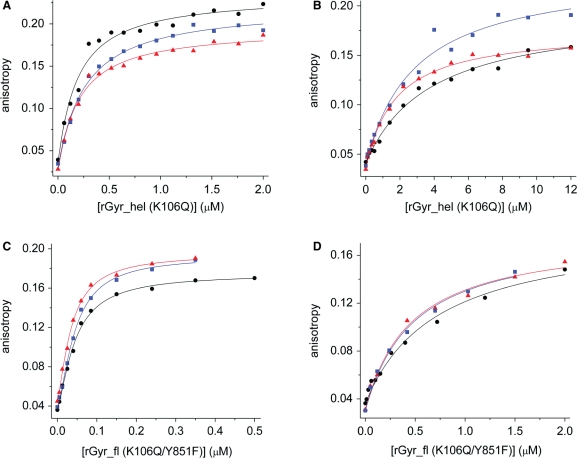
The DNA binding properties of this ATP-hydrolysis- and DNA-cleavage-deficient mutant were studied using the 60-base ssDNA and 60 bp dsDNA as a substrate (Figure 8, Table 5). ssDNA was bound independent of the nucleotide state, with Kd values of 44 ± 3 nM (no nucleotide), 44 ± 2 nM (ADP state) and 35 ± 2 nM (ATP state). Kd values for the corresponding dsDNA complexes were higher, consistent with the lower affinity of rGyr_fl for dsDNA. Similar to ssDNA binding, interaction of rGyr_fl(K106Q/Y851F) with dsDNA is independent of the nucleotide state. The Kd values of the dsDNA complexes are 0.71 ± 0.11 μM (no nucleotide), 0.46 ± 0.07 μM (ADP state) and 0.49 ± 0.09 μM (ATP state). Consistent with the wild-type data, the hydrolysis-deficient mutant of rGyr_fl thus shows ∼10-fold tighter binding of ssDNA to all nucleotide states.
Figure 8.
Binding of ssDNA and dsDNA to rGyr_hel(K106Q) and rGyr_fl(K106Q/Y851F). Titrations of ssDNA (A, C) and dsDNA (B, D) with rGyr_hel(K106Q) (A, B) or rGyr_fl(K106Q/Y851F) (C, D) in the absence of nucleotide (black, circles), and in the presence of ADP (2 mM, blue, squares) or ATP (4 mM, red, triangles). The DNA concentration was 25 nM in (A and B), and 10 nM in (C and D). The Kd values are summarized in Table 5.
The data for the wild-type enzyme and nonhydrolyzable ATP analogs (Figure 7, Table 5) support a slightly decreased preference for ssDNA after hydrolysis. The increased preference for ssDNA in the ADP state of rGyr_fl(K106Q/Y851F) results from a 2-fold increased Kd of 0.46 μM for the dsDNA complex, compared to a Kd of 0.25 μM for rGyr_fl(Y851F), and suggests differences between the ADP state of wild-type rGyr_fl and the K106Q/Y851F mutant. Previous work already pointed towards such a difference: Reverse gyrase relaxes negatively supercoiled DNA in the presence of ADP, whereas the K106Q mutant lacks such a relaxation activity (14). Most likely, lysine 106 functions as a nucleotide sensor that triggers an ADP-dependent conformational change that is required for relaxation. If this conformational change is absent in the K106Q mutant, the ADP state will be different, leading to different DNA binding properties.
To compare the DNA binding properties of the nucleotide states of rGyr_hel with rGyr_fl, the corresponding K106Q mutation in the Walker A motif of rGyr_hel was introduced, and the absence of hydrolysis was confirmed (data not shown). Fluorescence equilibrium titrations revealed a Kd value for the mantADP complex of 67 ± 5 μM, and 150 ± 85 μM and 570 ± 300 μM for the ADP and ATP complexes, respectively (Table 1). Thus, the nucleotide affinity of the K106Q mutant of rGyr_hel is ∼60-fold reduced compared to the wild-type. This effect is more pronounced than for rGyr_fl (20-fold).
rGyr_hel(K106Q) binds ssDNA 20-fold more tightly than dsDNA (Kd = 0.18 ± 0.03 and 3.8 ± 0.6 μM, respectively). In the presence of ADP or ATP, ssDNA binding is not significantly affected, and dsDNA binding is only slightly increased (ADP: 1.3-fold, Kd = 2.9 ± 0.6 μM; ATP: 2.2-fold, Kd = 1.7 ± 0.2 μM). As a consequence, ssDNA interacts more tightly than dsDNA with both nucleotide states (12-fold, ADP state; 9-fold, ATP state), implying a slightly stronger preference for ssDNA after ATP hydrolysis. Similarly, the data from wild-type rGyr_hel suggest a stronger interaction with ssDNA after ATP hydrolysis. However, wild-type rGyr_hel in complex with nonhydrolyzable ATP analogs binds ssDNA and dsDNA with similar affinities, while the ATP state of rGyr_hel(K106Q) interacts 9-fold more tightly with ssDNA. This discrepancy arises from a reduced affinity of the rGyr_hel(K106Q) ATP state for dsDNA compared to the ADPNP or ATPγS state of the wild-type rGyr_hel (Figure 6, Table 4), suggesting structural differences between the ATP state of rGyr_hel(K106Q) and the corresponding complexes of wild-type with nonhydrolyzable analogs.
Altogether, rGyr_hel shows similar affinities for ssDNA and dsDNA in the ATP state, and a 10-fold preference for ssDNA in the ADP state due to a decrease in dsDNA affinity. Thus, the helicase domain switches from a ‘high affinity’ state with respect to dsDNA to a ‘low affinity’ state upon ATP hydrolysis, and back to the ‘high affinity’ state after nucleotide exchange, while the ssDNA affinity remains constant. In rGyr_fl, both nucleotide states show a preference for ssDNA, indicating that the switch to the ‘low affinity’ state for dsDNA upon ATP hydrolysis is suppressed in the full-length enzyme. Figure 9 summarizes the properties of rGyr_hel and rGyr_fl. The K106Q mutants of rGyr_hel and rGyr_fl show similar DNA preferences, with a 10-fold higher affinity for ssDNA compared to dsDNA in both nucleotide states. These results argue for a critical role of lysine 106 as a nucleotide sensor in the helicase domain, and for inter-domain communication between the helicase-like and the topoisomerase domains.
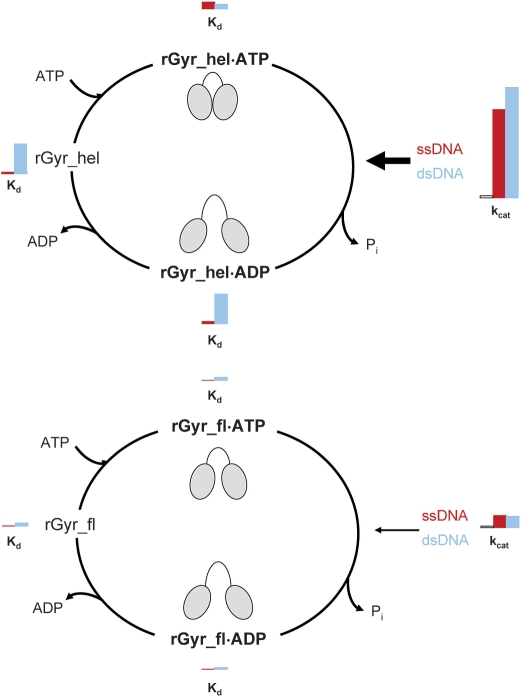
Figure 9.
Overview of the nucleotide cycle of rGyr_hel and rGyr_fl. The Kd values of the nucleotide states for ssDNA (red) and dsDNA (blue) are displayed as bars with a size proportional to their value. Analogously, a red bar denotes the kcat value in the presence of ssDNA, a blue bar corresponds to the kcat value in the presence of dsDNA. For reference, an additional bar represents the intrinsic kcat in the absence of DNA. A cartoon indicates possible conformations of the helicase-like domain in the different nucleotide states of rGyr_hel and rGyr_fl. rGyr_hel (upper panel) shows similar affinities for ssDNA and dsDNA in the ATP state, and a preference for ssDNA in the ADP state due to a decrease in dsDNA affinity. This corresponds to a switch from a ‘high affinity’ state with respect to dsDNA to a ‘low affinity’ state upon ATP hydrolysis. Both dsDNA and ssDNA efficiently stimulate ATP hydrolysis (arrow). Most likely, the helicase-like domain switches between a closed conformation (cartoon) in the presence of DNA and ATP that hydrolyzes ATP efficiently, and an open conformation in the ADP state, as observed for SF2 helicases. In rGyr_fl (lower panel), both nucleotide states show a preference for ssDNA, indicating that the switch to the ‘low affinity’ state for dsDNA upon ATP hydrolysis is suppressed in the full-length enzyme. The ATPase stimulation by DNA is 10-fold smaller than in rGyr_hel. Possibly, steric hindrance by the topoisomerase domain prevents a complete closure of the helicase-like domain (cartoon) in the context of full-length reverse gyrase. The progression through the nucleotide cycle with moderate velocity may be required for efficient coupling of ATP hydrolysis and positive supercoiling.
DISCUSSION
The helicase-like domain confers nucleotide-dependent DNA binding to reverse gyrase
We have shown here that the nucleotide binding properties of reverse gyrase are a function of the N-terminal helicase-like domain. Nucleotide affinities and ATP hydrolysis rates are virtually identical for the isolated helicase-like domain and for the full-length enzyme in the absence of nucleic acids. Thus, all determinants for nucleotide binding and hydrolysis are contained within the helicase-like domain, and its characteristics are unaltered in the context of reverse gyrase. The similar ATPase rates suggest that the conformations of the helicase-like domains are similar, indicating that the arrangement of the catalytic residues in the isolated helicase-like domain and in context of reverse gyrase are the same.
Our results demonstrate that the helicase-like domain mediates nucleotide-dependent interactions of reverse gyrase with ssDNA and dsDNA, and with plasmid DNA. In the ATP state, dsDNA and ssDNA are bound with similar affinities, whereas after ATP hydrolysis, ssDNA is preferentially bound. The DNA substrates influence the rate of switching between these states by stimulating the intrinsic ATPase activity of the helicase-like domain. Thus, the helicase-like domain is a module that confers nucleotide-dependent DNA binding to reverse gyrase. The topoisomerase domain has no influence on nucleotide binding and hydrolysis by the helicase-like domain in the absence of DNA, but modulates the ATPase properties in the presence of DNA. In the context of reverse gyrase, the ATPase activity of the helicase-like domain is still stimulated by the same DNA substrates, but to a much lesser extent. The DNA-stimulated ATPase activity of reverse gyrase is 10-fold lower than that in the isolated domain, demonstrating that the activity of the helicase-like domain is strongly suppressed by the topoisomerase domain.
The nucleic acid-stimulated ATPase activity of SF2 helicases results from a conformational change upon binding of both ligands that leads to a closure of the cleft between the two helicase subdomains. In this closed conformation, the catalytic residues are correctly positioned for efficient ATP hydrolysis. It has been proposed that such a conformational change in the helicase-like domain initiates supercoiling by reverse gyrase (10,14). The observed similar ATPase activities of the helicase-like domain and reverse gyrase suggest a similar open conformation of the helicase in both enzymes (Figure 9). In the presence of DNA, the isolated helicase-like domain adopts a closed conformation that rapidly hydrolyzes ATP. Possibly, in full-length reverse gyrase the topoisomerase domain provides a steric hindrance for the conformational change in the helicase-like domain. This steric hindrance may confine the helicase-like domain to a more open form even in the presence of nucleic acids, and consequently to less efficient ATP hydrolysis by reverse gyrase compared to the isolated helicase-like domain. A restriction of this conformational change by the topoisomerase domain would also readily explain the lower cooperativity between ATP and nucleotide binding in rGyr_fl.
Inter-domain communication in reverse gyrase: DNA binding and ATPase stimulation
The helicase-like domain and reverse gyrase interact more tightly with ssDNA than with dsDNA. This preference for ssDNA is reminiscent of other type I DNA topoisomerases (27). The high affinity of reverse gyrase for ssDNA may allow for sensing of single-stranded regions (5), and lead to stabilization of these regions for strand cleavage and the subsequent strand-passage reaction during catalysis. It has been shown previously that the presence of single-stranded regions in the DNA substrate favors positive supercoiling by reverse gyrase (5). Reverse gyrase contains several potential interaction sites for nucleic acids, located in the helicase-like domain, or in a cleft close to the catalytic tyrosine. In addition, the latch region may contact DNA (10,12), and a transient interaction of the putative zinc fingers with the DNA during supercoiling has been proposed (10). However, nucleic acid binding to these different interaction sites of reverse gyrase has not been characterized in detail. By using the Y851F mutant of reverse gyrase in DNA binding studies, we excluded the covalent binding of DNA via the catalytic tyrosine. In general, the DNA affinity of reverse gyrase is about 10-fold higher than that for the helicase-like domain. This increased affinity could either reflect tight DNA binding to the helicase-like domain in reverse gyrase, or point towards simultaneous interactions of DNA with the topoisomerase domain that increase the overall affinity. Clearly, a more detailed analysis of DNA binding to reverse gyrase would be required to further understand the relation between DNA binding to different sites and stimulation of ATP hydrolysis. However, in this context it is important to note that the cooperativity observed for ssDNA binding does not reflect two DNA molecules binding to one molecule of reverse gyrase (as would be possible if one ssDNA molecule interacts with the helicase-like domain and one with the topoisomerase domain). Instead, the data is consistent with the interaction of two reverse gyrase molecules with one ssDNA molecule, pointing towards a possible interaction between reverse gyrase molecules on the same substrate DNA. The fact that this cooperative effect is more pronounced for the full-length enzyme than for the isolated helicase-like domain, and only observed for ssDNA but not for dsDNA, suggests a functional role of this protein–protein interaction during the supercoiling reaction.
Is the ATPase activity of the helicase-like domain modulated to achieve optimal coupling of ATP hydrolysis to positive supercoiling?
Our results demonstrate that the helicase-like domain of reverse gyrase meets most criteria for a bona fide helicase: it has a DNA-dependent ATPase activity, and the nucleotide-state determines its affinity for ssDNA and dsDNA. However, as already reported for other reverse gyrase homologs, we did not detect DNA unwinding activity for the helicase-like domain of T. maritima reverse gyrase (data not shown). Altogether, the helicase-like domain of reverse gyrase acts as a nucleotide-dependent switch reminiscent of the nonprocessive DEAD box helicases that constitute a subfamily of SF2 helicases (28).
Helicase domains frequently occur in conjunction with additional domains that mediate substrate specificity or the interaction with protein partners, or regulate helicase activity (29–33). For instance, the DEAD box helicase RIG-I consists of a helicase domain and two N-terminal caspase activation and recruitment domains that connect the helicase function to the caspase signaling pathway, and these additional domains inhibit the RIG-I helicase activity (33). Conversely, the internal helicase domain of dicer inhibits its RNase activity (32), demonstrating that the domains can mutually influence their respective activities. Likewise, the ATPase activity of the helicase-like domain in the presence of DNA is attenuated in the context of reverse gyrase, and the differences in nucleic acid affinities of the nucleotide states are smaller (Figure 9). A repression of relaxation activity of the topoisomerase domain in the absence of nucleotides by the helicase-like domain, mediated by the latch region (11,12), has been demonstrated earlier. The inhibition of the helicase-like domain we describe here seems to be the reciprocal effect. Deletion of the latch region increases the ATPase activity of reverse gyrase in the presence of ssDNA (11), suggesting a simultaneous role of the latch for the inhibitory effect of the topoisomerase domain on the helicase-like domain. Both effects emphasize the importance of communication between the reverse gyrase domains. The fact that the differences between switching of the helicase-like domain and reverse gyrase are mostly lost in the K106Q mutants suggests a critical contribution of the Walker A motif to inter-domain communication.
As a consequence of the inhibitory effect of the topoisomerase domain on the helicase-like domain in reverse gyrase, the progression through the nucleotide cycle in the presence of DNA is strongly decelerated. DNA supercoiling is a complex multi-step process, and the current model of DNA supercoiling by type IA topoisomerases such as reverse gyrase involves DNA cleavage, strand passage and re-ligation of the cleaved strand. This reaction will most likely require substantial conformational changes in reverse gyrase, and supercoiling is therefore an intrinsically slow process. Thus, a rapid cycling between the nucleotide states may not be necessary or could even be detrimental for supercoiling. Instead, the precise coordination of ATP hydrolysis and supercoiling may require matching rates for both processes, and hence a moderate ATP turnover. The inhibitory effect of the topoisomerase domain on the helicase-like domain emphasizes the role of inter-domain communication for efficient coupling between ATP hydrolysis and positive DNA supercoiling by reverse gyrase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
VolkswagenStiftung; Swiss National Science Foundation. Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Manuel Hilbert for performing the unwinding experiment with rGyr_hel and Markus Rudolph for discussions.
REFERENCES
- 1.Kikuchi A, Asai K. Reverse gyrase–a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984;309:677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- 2.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 3.Atomi H, Matsumi R, Imanaka T. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 2004;186:4829–4833. doi: 10.1128/JB.186.14.4829-4833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kampmann M, Stock D. Reverse gyrase has heat-protective DNA chaperone activity independent of supercoiling. Nucleic Acids Res. 2004;32:3537–3545. doi: 10.1093/nar/gkh683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh TS, Plank JL. Reverse gyrase functions as a DNA renaturase: annealing of complementary single-stranded circles and positive supercoiling of a bubble substrate. J. Biol. Chem. 2006;281:5640–5647. doi: 10.1074/jbc.M513252200. [DOI] [PubMed] [Google Scholar]
- 6.Krah R, Kozyavkin SA, Slesarev AI, Gellert M. A two-subunit type I DNA topoisomerase (reverse gyrase) from an extreme hyperthermophile. Proc. Natl Acad. Sci. USA. 1996;93:106–110. doi: 10.1073/pnas.93.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krah R, O’Dea MH, Gellert M. Reverse gyrase from Methanopyrus kandleri. Reconstitution of an active extremozyme from its two recombinant subunits. J. Biol. Chem. 1997;272:13986–13990. doi: 10.1074/jbc.272.21.13986. [DOI] [PubMed] [Google Scholar]
- 8.Declais AC, Marsault J, Confalonieri F, de La Tour CB, Duguet M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J. Biol. Chem. 2000;275:19498–19504. doi: 10.1074/jbc.M910091199. [DOI] [PubMed] [Google Scholar]
- 9.Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc. Natl Acad. Sci. USA. 1993;90:4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez AC, Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez AC. Studies of a positive supercoiling machine. Nucleotide hydrolysis and a multifunctional “latch” in the mechanism of reverse gyrase. J. Biol. Chem. 2002;277:29865–29873. doi: 10.1074/jbc.M202853200. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez AC. Investigating the role of the latch in the positive supercoiling mechanism of reverse gyrase. Biochemistry. 2003;42:5993–6004. doi: 10.1021/bi034188l. [DOI] [PubMed] [Google Scholar]
- 13.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl Acad. Sci. USA. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jungblut SP, Klostermeier D. Adenosine 5'-O-(3-thio)triphosphate (ATPgammaS) promotes positive supercoiling of DNA by T. maritima reverse gyrase. J. Mol. Biol. 2007;371:197–209. doi: 10.1016/j.jmb.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 15.Peck ML, Herschlag D. Adenosine 5'-O-(3-thio)triphosphate (ATPgammaS) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A. RNA. 2003;9:1180–1187. doi: 10.1261/rna.2103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Kovalsky OI, Kozyavkin SA, Slesarev AI. Archaebacterial reverse gyrase cleavage-site specificity is similar to that of eubacterial DNA topoisomerases I. Nucleic Acids Res. 1990;18:2801–2805. doi: 10.1093/nar/18.9.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaxel C, Duguet M, Nadal M. Analysis of DNA cleavage by reverse gyrase from Sulfolobus shibatae B12. Eur. J. Biochem. 1999;260:103–111. doi: 10.1046/j.1432-1327.1999.00128.x. [DOI] [PubMed] [Google Scholar]
- 19.Hiratsuka T. New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim. Biophys. Acta. 1983;742:496–508. doi: 10.1016/0167-4838(83)90267-4. [DOI] [PubMed] [Google Scholar]
- 20.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 21.Shibata T, Nakasu S, Yasui K, Kikuchi A. Intrinsic DNA-dependent ATPase activity of reverse gyrase. J. Biol. Chem. 1987;262:10419–10421. [PubMed] [Google Scholar]
- 22.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 23.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 24.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 25.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 26.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 27.Kirkegaard K, Wang JC. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 1985;185:625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- 28.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 29.Morlang S, Weglohner W, Franceschi F. Hera from Thermus thermophilus: the first thermostable DEAD-box helicase with an RNase P protein motif. J. Mol. Biol. 1999;294:795–805. doi: 10.1006/jmbi.1999.3282. [DOI] [PubMed] [Google Scholar]
- 30.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 31.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 32.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gee P, Chua PK, Gevorkyan J, Klumpp K, Najera I, Swinney DC, Deval J. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J. Biol. Chem. 2008;283:9488–9496. doi: 10.1074/jbc.M706777200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.