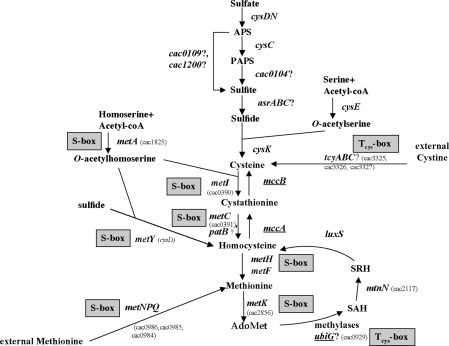
Figure 1.
Sulfur metabolism in C. acetobutylicum. The genes controlled by antisense RNAs are underlined. ‘S-box’ indicates the genes controlled by an S-box riboswitch. And ‘Tcys-box’ indicates the genes controlled by a cysteine-specific T-box system. A question mark indicates the genes probably involved in this pathway. AdoMet, S-adenosyl-methionine; Serine O-acetyltransferase, cysE; OAS-thiol-lyase, cysK; methionine adenosyltransferase, metK; adenosylhomocysteine nucleosidase, mtnN; S-ribosylhomocysteine lyase, luxS; cystathionine β-synthase, mccA; cystathionine γ-lyase, mccB; homoserine acetyl-transferase, metA; cystathionine γ-synthase, metI; cystathionine β-lyase, metC and patB; methionine synthase, metH; ATP sulfurylase, cysDN; APS kinase, cysC; anaerobic sulfite reductase, asrABC (11,12). The cac numbers for C. acetobutylicum genes correspond to those of AcetoList (http://bioinfo.hku.hk/GenoList/index.pl?database=acetolist) (38).