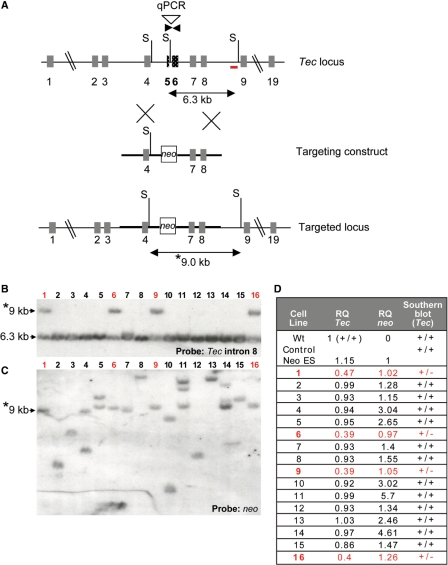
Figure 1.
Screening by Southern blot and q-PCR of ES clones targeted with a conventional construct. (A) Schematic showing Tec targeting vector designed to delete 1.2 kb including exons 5 and 6. The primers used for q-PCR are indicated in dark inverse arrows. The 3′ Tec probe for Southern blotting is indicated as the red bar (in intron 8). S: SspI sites in the construct and the endogenous gene. Double arrows indicated the endogenous and targeted SspI fragments detected in Southern analyses. (B, C) Southern blot analysis of ES DNA digested with SspI and hybridized with (B) the 3′ probe (located in intron 8 of the Tec gene). The migration of the endogenous 6.3 kb SspI band and the targeted 9.0 kb fragment (asterisk) are indicated. (C) The same membrane was stripped and re-hybridized with a neo probe. The targeted 9.0 fragment (asterisk) is indicated. (D) q-PCR was performed using ES DNA from clones previously examined by Southern blotting and primers for the deleted region of Tec, neo and a reference gene Itk. As calibrators, DNA from WT ES cells (containing two copies of the deleted Tec region) and from an ES cell clone harboring one copy of neo were used with RQ values set to 1. Results are compared to Southern analyses. Targeted clones have RQ values close to 0.5 for Tec and 1 for neo. Clones containing a disruption of Tec are indicated in red.