Abstract
The loxP/Cre recombination system is a widely used tool for mouse functional genomics, in particular for in vivo conditional mutagenesis. Depending on the relative orientation and position of loxP sites, Cre-mediated recombination can result in a variety of targeted genomic rearrangements. It was previously reported that loss of the loxP-carrying chromosome can occur when loxP sites are arranged in inverse orientation. By using a chromosome 2 carrying inverted loxP sites, we found that Cre-mediated recombination not only causes chromosomal loss but also triggers apoptosis. We show that targeted recombination between inverted loxP sites (TRIP) triggers cell death specifically in proliferating Cre-expressing cells, and we provide evidence that TRIP is an efficient tool to ablate proliferating cells within genetically defined cell populations. Furthermore, the procedure requires only a simple, one-step intercross but neither the use of toxins nor the additional step of prodrug injection. With the large repertoire of tissue-specific or inducible Cre-expressing transgenes available, TRIP-mediated cell ablation is valuable to investigate the function of a large variety of cell populations in the context of a whole organism, which includes mechanisms underlying organ development and tissue homeostasis.
Keywords: apoptosis, chromosome loss, Cre recombinase, genetic ablation
The site-specific recombinase activity of the bacteriophage P1 Cre enzyme is a highly accurate recombination system that has become a major tool for functional genomics in mice. A variety of targeted genomic rearrangements can be generated depending on the relative position and orientation of the Cre target sequence (loxP site). These rearrangements include targeted deletions, inversions, and duplications (1, 2). Importantly, the loxP/Cre-mediated recombination has provided a means to circumvent drawbacks associated with ubiquitous gene inactivation or gain-of-function experiments, through the development of inducible or tissue-specific Cre transgenes (3). In this respect, the use of the loxP/Cre system has had a considerable impact on the study of gene function.
In most cases, loxP/Cre-mediated recombinations generate viable rearrangements. However, when loxP sites located in cis are in inverse orientation with respect to each other, recombination can result in the elimination of the loxP-carrying chromosome (1, 4, 5). Lewandoski et al. showed that combining a Y chromosome carrying loxP sites in inverted orientation with a Cre transgene expressed ubiquitously during early embryogenesis resulted in XX and XO progeny, which indicated that the Y chromosome had been eliminated (4). Based on their findings, they proposed that recombination between two loxP sites in inverted orientation could be used as a tool to target chromosome loss. More recently, inverted loxP sites were used to target the elimination of embryonic stem (ES) cell-derived chromosomes in tetraploid ES-somatic hybrids (5).
In the present study, we investigated the possibility of generating tissue-restricted monosomies by inducing targeted recombination between inverted loxP sites (referred to as TRIP hereafter) in a tissue-specific manner. For this purpose, we produced double heterozygous embryos carrying a set of inverted loxP sites on chromosome 2 and Cre transgenes expressing the recombinase in distinct cell populations. Consistent with previous reports (4, 5), targeted recombination between inverted loxPs (TRIP) resulted in chromosome loss in proliferating cells. Unexpectedly, we also found that cells that have lost the loxP-carrying chromosome were eliminated by apoptosis before completion of the cell cycle, which indicated that the recombination outcome was cytotoxic. The extent of cell death within the Cre-positive domain indicated that this phenomenon occurred with a high incidence, and we provide evidence that TRIP is an efficient and simple genetic means to perform specific ablation of proliferating cells. The possibility to block the expansion of genetically defined cell populations through TRIP-mediated elimination of proliferating cells offers a valuable tool to study both morphogenetic processes and mechanisms underlying tissue/organ homeostasis and regeneration.
Results
Recombination Between loxP Sites with Inverse Orientation Induces Apoptosis.
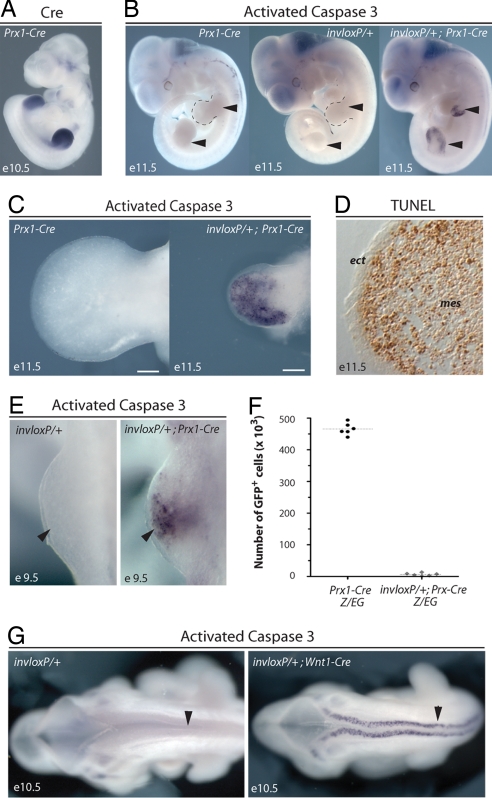
Recombination between loxP sites in inverted orientation has been proposed as a tool to induce a targeted loss of chromosome and monosomies in a tissue-specific manner (4), which would circumvent the embryonic lethality associated with constitutive autosomal monosomies (6). We used this approach to generate a tissue-specific monosomy of chromosome 2 (Chr2). We crossed mice having a set of loxP sites in inverted orientation within the 5′ part of the HoxD gene cluster (referred to as invloxP hereafter) (7) to mice carrying a Cre transgene expressed primarily in developing limbs (Prx1-Cre) (8), as shown in Fig. 1A. We found that limb buds of invloxP/+; Prx1-Cre embryos were severely reduced in size as compared with wild-type ones. In contrast, embryos carrying only the invloxP allele or the Prx1-Cre transgene were indistinguishable from wild-type embryos (Fig. 1 B and C) and were used as controls in subsequent analyses. Immunodetection of the activated form of caspase 3 and TUNEL assays revealed that there was massive apoptosis in the mesenchyme of invloxP/+; Prx1-Cre limb buds (Fig. 1 B–D). In contrast, adjacent ectodermal cells, which did not express the Cre recombinase, were not affected (Fig. 1D), which established that the induced ectopic cell death was restricted to Prx1-Cre-expressing cells. Apoptosis was detected already in nascent limb buds, indicating that ectopic cell death began soon after the expression of the Cre recombinase (Fig. 1E).
Fig. 1.
Targeted recombination between inverted loxP sites (TRIP) induces apoptosis. (A) Cre mRNA detected by whole-mount in situ hybridization in e10.5 Prx1-Cre embryo. (B) Whole-mount immunodetection of the activated form of caspase 3 showing that massive apoptosis is detected in invloxP/+; Prx1-Cre limb buds (Right; arrowheads) but not in controls (Left and Center; arrowheads). (C) Higher magnification of control and mutant forelimb buds at e11.5. (D) TUNEL assay on cryosection of mutant forelimb bud. Induced apoptosis is detected in mesenchymal cells but not in ectodermal cells. (E) Whole-mount immunodetection of activated caspase 3 in nascent mutant forelimb buds (e9.5). (F) Number of GFP positive cells (i.e., Prx1 lineage, see Materials and Methods) per forelimb bud isolated from control and mutant embryos at 49–50 somites stage. Number of cells and mean value for at least six different limb buds per genotype are shown. Between 1,000 and 13,000 GFP-positive cells remained per mutant limb bud. (G) TRIP-induced apoptosis is detected in Cre-expressing cells of invloxP/+; Wnt1-Cre embryos (Right; arrowhead). mes, mesenchyme, ect, ectoderm. invloxP, Chromosome 2 carrying inverted loxP sites. (Scale bar: 200 μm.)
TRIP Results in the Depletion of Genetically Defined Cell Populations.
Our analyses revealed that TRIP resulted in widespread cell death within the Cre-expression domain. To assess the severity of this effect, the Z/EG reporter transgene, permanently expressing green fluorescent protein (GFP) in Cre-expressing cells and all their progeny (9) was used to label the Prx1-Cre lineage in both control (Z/EG; Prx1-Cre) and mutant (invloxP/+; Z/EG; Prx1-Cre) limb buds. We dissected out limb buds from control and mutant embryos at the 49 to 50 somite stage and determined the total number of cells expressing the GFP. Between 1,000 and 13,000 GFP-positive cells remained per mutant limb bud, whereas control buds contained ≈470,000 GFP-positive cells (Fig. 1F), indicating that 48 h after the onset of Cre expression less than 3% of the Prx1-Cre cell lineage remained.
The DNA fragment flanked by the two loxP sites contained two genes, Hoxd12 and Hoxd11. Limb buds of heterozygous embryos carrying either the inversion (7) or deletion (10) of this piece of DNA were morphologically indistinguishable from wild-type buds. Therefore, we concluded that the ectopic apoptosis induced following Cre-mediated recombination was not due to impaired function of HoxD genes. To further confirm that cell death was unrelated to the function of HoxD genes, we investigated the effect of combining the invloxP allele with the Wnt1-Cre transgene, which is expressed in neural crest cells (11). Analysis of invloxP/+; Wnt1-Cre embryos showed massive apoptosis in the entire Wnt1-Cre expression domains (Fig. 1G) including in embryonic regions that never expressed 5′ HoxD genes. Here again, embryos carrying only the Cre transgene or the inverted loxP sites were indistinguishable from wild-type embryos (Fig. 1G and data not shown).
Together, these results suggested that Cre-mediated recombination between loxP sites in opposite orientation triggered apoptosis and resulted in the specific depletion of Cre-expressing cells.
Loss of the loxP-Carrying Chromosome Is Detrimental for Cell Survival.
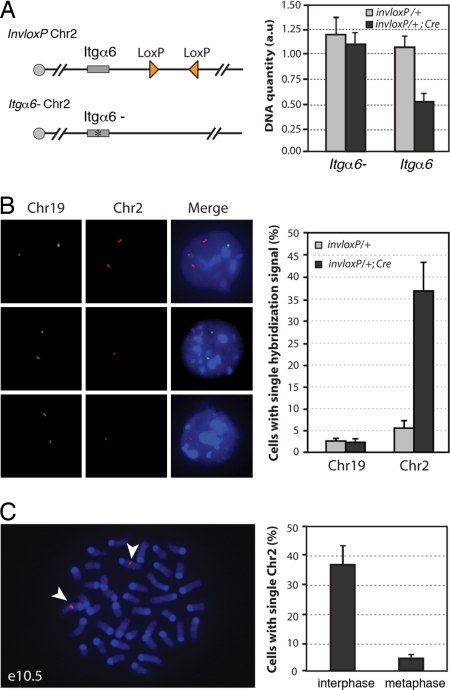
Because recombination between cis-located loxP sites in inverse orientation could result in the elimination of the loxP-carrying chromosome (1, 4), we investigated whether the induction of ectopic cell death was associated with chromosome loss. For this purpose, we first performed chromosome quantification in limb bud cells. To distinguish the loxP-carrying Chr2 from its homologous counterpart, we produced embryos with one copy of Chr2 carrying the invloxP allele and the wild-type allele of integrin alpha 6 (Itgα6) and the other copy of Chr2 carrying a mutant allele of Itgα6 (12), with or without the Cre transgene (Fig. 2A). Quantification by real-time PCR of the wild-type versus mutant Itgα6 allele provided a means to establish the relative proportion of the loxP-carrying Chr2 versus its homologue. We found that the wild-type Itgα6 allele was almost twofold underrepresented in invloxP/Itgα6-; Prx1-Cre limb bud cells as compared with invloxP/Itgα6- (Fig. 2A, right panel), indicating that, in invloxP/+; Prx1-Cre embryos, the Chr2 carrying inverted loxPs was missing in about half of the limb bud cells.
Fig. 2.
TRIP results in chromosome loss and cell death in proliferating cells. (A) Real-time PCR detection of loss of the loxP-carrying Chr2. Chr 2 carrying inverted loxP sites (triangles) was combined with a Chr2 with a mutation in the integrin α6 gene (Itgα6-) (see Materials and Methods). Integrin α6 quantification values were normalized using the Hoxa13 gene located on Chr6. Data are presented as means and ranges from two independent experiments. (B) FISH detection of Chr2 monosomy. In e10.5 invloxP/+; Prx1-Cre limb buds, some cells showed two hybridization signals for Chr2 (Left, top row) while others had a single hybridization signal (Left, middle and bottom rows). For each experiment, at least 200 cells for each genotype were analyzed. Data are presented as means ± SD from three independent experiments. Background level of cells with single signal for Chr19 or Chr2 (3–6%) most likely corresponds to technical limitation or FISH signals in close vicinity to each other (see Materials and Methods) (C) FISH detection of Chr2 in metaphase cells from invloxP/+; Prx1-Cre forelimb buds at e10.5. More than 50 metaphases were analyzed. Representative metaphase with two copies of Chr2 (arrowheads). Data are presented as means ± SD from three independent experiments. Green, DIG probe; red, biotin probe.
To independently assay for chromosome loss, we performed fluorescent in situ hybridization (FISH) for Chr2 on dissociated cells from invloxP/+ (control) and invloxP/+: Prx1-Cre (mutant) forelimb buds. FISH for Chr19 was used as internal control. Among live cells isolated from mutant buds, 37% (±5%) had a single hybridization signal with Chr2-specific probes (Fig. 2B). In marked contrast, we repeatedly detected both copies of Chr2 in metaphasic cells (Fig. 2C). This result indicated that cells that have lost the loxP-carrying chromosome have been eliminated by apoptosis before entering in metaphase. The striking absence of cells with a Chr2 monosomy in M phase, in contrast to the high incidence of monosomy in interphase (more than one-third of cells contained a single Chr2), suggested that cell death had occurred as a consequence of chromosome elimination.
TRIP-Mediated Cell Death Occurs Specifically in Proliferating Cells.
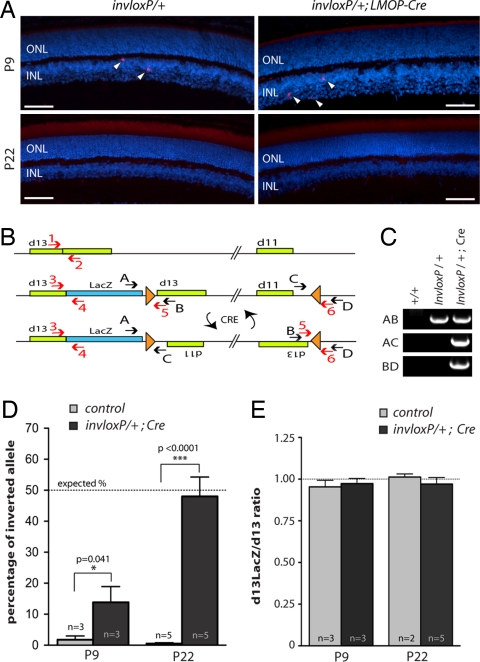
The elimination of a chromosome as a consequence of Cre-mediated recombination between inverted loxP sites was proposed to be the result of unequal crossover between sister chromatids, after DNA replication and before entry into anaphase (4). Our observations that cells that have lost one copy of their Chr2 were eliminated before completion of the cell cycle suggested that TRIP-mediated cell death was associated with chromosome loss and might therefore be restricted to cycling cells. To test whether the induction of apoptosis was specific to proliferating cells, we examined the effects of TRIP on postmitotic cells. We generated mutants carrying the invloxP allele together with the LMOP-Cre transgene, which is specifically expressed in postmitotic photoreceptor cells (13). Retinas of invloxP/+; LMOP-Cre mice were indistinguishable from controls. TUNEL assay and immunodetection of the activated form of caspase 3 on retinas of 9 days (P9) and 22 days (P22) invloxP/+; LMOP-Cre and control mice did not reveal any ectopic cell death in mutant retinas (Fig. 3A), suggesting that Cre activity in postmitotic photoreceptors did not induce cell death. Nevertheless, we were able to detect inversion of the DNA fragment located in between both inverted loxP sites (Fig. 3 B and C) indicating that recombination had occurred but did not affect cell survival. A possibility existed, however, that Cre-mediated recombination in postmitotic photoreceptors had triggered cell death but was barely detectable due to ineffective Cre-mediated recombination. Although this latter possibility was unlikely based on the characterization of the LMOP-Cre transgene previously reported (13), we verified the efficiency of LMOP-Cre-mediated recombination. For this purpose, we quantified by real-time PCR the cell population carrying the inversion of the DNA fragment flanked with loxP sites. The inversion event being reversible because of the maintenance of the two loxP sites after recombination, an average of 50% of Cre-expressing cells was expected to carry the inverted DNA fragment at a given time point. Quantification of the inverted fragment showed that, whereas only 14% of photoreceptors carried the inverted allele at P9 (instead of the 50% expected), the inverted DNA fragment was present in 48% of the photoreceptor population at P22 (Fig. 3D). These results indicated that LMOP-Cre triggered recombination only in a quarter of photoreceptors at early stages of Cre expression but recombination extended to almost the entire population by P22. Nevertheless, we did not observe ectopic cell death at these stages indicating that TRIP did not trigger apoptosis in postmitotic cells. Furthermore, there was no chromosome loss detectable (Fig. 3E), which confirmed the link between chromosome loss and the induction of apoptosis. Together, these results indicated that TRIP triggered apoptosis specifically in proliferating cells.
Fig. 3.
TRIP-mediated chromosome loss and cell death is specific to proliferating cells. (A) Immunodetection of the activated form of caspase 3 on cryosections of invloxP/+ and invloxP/+;LMOP-Cre retinas at P9 and P22. Nuclei were counterstained with DAPI. Some apoptotic cells (arrowheads) were detected in the inner nuclear layer (INL) at P9. No difference was detected between control and mutant retinas. (B) Scheme of wild-type, noninverted, and inverted loxP-carrying Chr2. LoxP sites (triangles) and primers used for PCR (black arrows) and real-time PCR (red arrows) are indicated. (C) PCR detection of DNA inversion in invloxP/+;LMOP-Cre retinas at P22. Cre-mediated inversion of the DNA fragment flanked with loxP sites is detected with the AC and BD primer sets. (D) Percentage of inverted allele (primer pair 5–6) in control and mutant retinas, at P9 and P22. If Cre is active in all photoreceptors, the percentage of inverted allele should be 50% on average, as the inversion is reversible (this theoretical percentage is indicated by a dotted line). (E) Quantification of loxP-carrying allele (primer pair 3 and 4) versus wild-type allele (primer pair 1 and 2) in control and mutant retinas at P9 and P22. Data are presented as means ± SEM. Quantification resulted from average of three independent quantitative PCR reactions. Number of animals for each type and P values from unpaired t test are indicated. ONL, outer nuclear layer; INL, inner nuclear layer. (Scale bar: 50 μm.)
Discussion
Recombination between inverted loxP sites was previously reported to result in the elimination of the loxP-carrying chromosome and was envisioned as a means to generate chromosome elimination in a targeted manner (4, 5). In this study, we used this approach to generate a tissue-specific monosomy of chromosome 2. However, we discovered that Cre-mediated recombination between inverted loxP sites triggered massive cell death within Cre expression domains. Further analysis revealed that the loss of chromosome 2 in proliferating cells induced apoptosis before the onset of mitosis, indicating that this genomic rearrangement was ultimately cytotoxic for proliferating cells. The extent of chromosome loss and subsequent death of proliferating Cre-expressing cells resulted in a severe depletion of the Cre cell lineage. In this respect, TRIP appears to be an efficient means to ablate proliferating cells within genetically defined cell populations. TRIP thus provides a versatile novel tool to assess morphogenetic processes underlying organ development, tissue homeostasis and regeneration.
The link between the loss of chromosome 2 and the activation of the apoptotic cascade is intriguing. Chromosome elimination occurs when TRIP takes place during the S or G2 phase of the cell cycle as a result of transrecombination between sister chromatids (1, 4). In contrast, during the G1 phase, recombination between inverted loxP sites occurs in cis, resulting in the inversion of the intervening DNA fragment that is not per se cytotoxic. Because both loxP sites are maintained after inversion, recombination in G1 does not preclude subsequent recombination during S or G2 phase. The extent of chromosome loss that we observed indicated that unequal exchanges between sister chromatids occurred with a high incidence. Cells with a chromosome 2 monosomy were detectable in interphase but were eliminated before entry into metaphase, indicating that apoptosis was induced soon after chromosome loss. Activation of the apoptotic pathway could be a consequence of the hemizygous status of chromosome 2 if this chromosome carries essential gene(s) or regulatory elements critical to cell survival. Alternatively, chromosome loss could be detected by the cell cycle checkpoint machinery, which would then trigger the apoptotic cascade to kill aneuploïd cells. In this view, the viability of cells in which loss of the Y chromosome was induced (4) suggests that TRIP-mediated cell death would be associated with cell-cycle checkpoint mechanism that does not monitor the Y-chromosome.
The activation of the apoptotic cascade following the loss of chromosome 2, and the likelihood that elimination of other autosomes could cause similar effects, imply that careful controls should be considered whenever using the loxP/Cre system. The recombination-specific, yet gene-independent, induction of cell death could confound interpretation of experimental data. For instance, upon pronuclear injection, transgene constructs rarely integrate as single copy and inverted insertion occurs frequently within a transgene array. Even if the effect is specific to chromosome 2, it is one of the largest murine chromosomes, and therefore there is a higher probability of transgene insertion within that specific chromosome.
In developing forelimb buds, 48 h after the onset of Cre expression, the Prx1-Cre cell lineage was reduced to less than 3% in comparison to control limb buds. Based on the extent of cell death observed, TRIP appears as an effective genetic tool to ablate proliferating Cre-expressing cells. Previously reported genetic methods for targeted cell ablation are primarily based on conditional expression of the diphtheria toxin (DTA) (14–16) or its receptor (17, 18), which target virtually all Cre-expressing cells irrespective of their cell cycle status. In contrast, TRIP triggers cell death specifically in proliferating cells. As a consequence, TRIP is a convenient means of interfering with the expansion of genetically defined cell populations and thereby can be useful in studying the role of cell proliferation during tissue/organ homeostasis and regeneration. For instance, whether in vivo homeostasis involves the contribution of putative stem cells or of differentiated cells is still an unresolved issue for a large number of systems, mostly due to the difficulty of tracing back the origin of cells implicated in these regulatory processes and the putative involvement of slow dividing cells (19). TRIP provides a simple tool for assaying distinct cell populations with respect to their ability to participate in organ maintenance and regeneration. Furthermore, TRIP-mediated cell ablation circumvent the experimental difficulties related to slow dividing cells as it offers the possibility to monitor the effect of the ablation of dividing cells over extensive time periods.
Genetic fate mapping has proved to be an insightful complement to functional genomics to investigate morphogenetic processes during embryogenesis. The possibility to specifically impede the expansion of a genetically defined cell population through targeted ablation of proliferating Cre-expressing cells offers a novel approach to further refine assessment of developmental processes. For instance, TRIP-mediated cell ablation is a straightforward means to investigate the extent of crosstalk between neighboring but genetically distinct cell populations. Combined with genetic fate mapping, it can also be used as a tool to assay cell fate specification versus determination during organogenesis. Furthermore, because TRIP-mediated ablation occurs in double heterozygous specimens, series of mutants are easily obtained for comprehensive analysis of cell ablation outcomes.
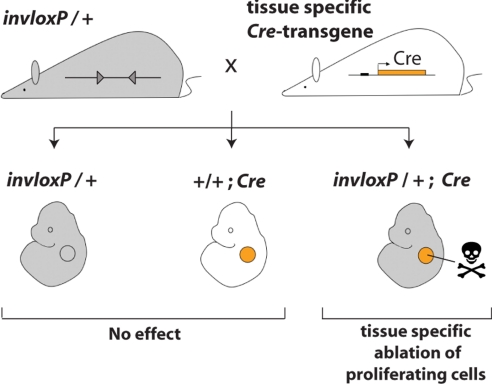
The TRIP procedure is fast and simple, as it involves only a single breeding step (Fig. 4). Of note, TRIP-mediated cell ablation does not require any injections to activate the system, and it thereby prevents problems of targeting embryonic tissues caused by the placental barrier and also limits variations caused by experimental manipulations. Furthermore, TRIP-based cell ablation circumvents potential secondary effects that can be associated to toxin-mediated cell ablation approaches, in particular when using the human heparin-binding epidermal growth factor as a receptor for the diphtheria toxin (20). In conclusion, TRIP is a versatile and simple tool for generating targeted ablation of genetically defined proliferating cell populations, which will be valuable for studying both morphogenetic processes and mechanisms underlying tissue/organ homeostasis and regeneration.
Fig. 4.
Experimental design for TRIP-mediated ablation of proliferating cells. The mouse strain carrying the chromosome 2 with loxP sites in inverse orientation (triangles, invloxP allele) is crossed with a mouse expressing a Cre transgene under the control of a tissue-specific promoter. In the resulting progeny, ablation of proliferating cells due to the recombination between the inverted loxP sites occurs specifically in Cre-expressing tissue (orange circle) of double heterozygous specimens (invloxP/+; Cre).
Materials and Methods
Animals.
Mouse lines used in this work were described previously: InvloxP (7), Prx1-Cre (8), Wnt1-Cre (11), LMOP-Cre (13), Z/EG (9). Genotyping was done by Southern blot analysis using genomic DNA isolated from tail biopsy samples or yolk sacs.
Apoptosis Detection.
Whole-mount immunodetection of cleaved caspase 3.
Embryos were treated with 5% H2O2 in methanol for 1 h, blocked in PBSMT (×1 PBS, 2% milk, 2.5% Triton X-100) for 1 h, and incubated with anti-caspase 3 antibody (Cell Signaling Technology, #9661) 1:100 in PBSMT, overnight at 4°C. After extensive washes for 5 h in PBSMT, embryos were incubated with AP-conjugated goat anti-rabbit (Santa Cruz Biotechnology) 1:2000 in PBSMT overnight at 4°C. Embryos were equilibrated in NTMT (100 mM Tris pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween-20) and alkaline phosphatase activity detected using NBT/BCIP substrate (Roche).
Apoptosis detection on cryosections.
Apoptotic cells were detected by immunodetection of cleaved caspase 3 (Cell Signaling Technology, #9661) or the deoxynucleotidyltransferase-mediated dUTP-biotin nick-end labeling assay (TUNEL, Promega) on cryosections of limb buds or retinas (14 μm) following classical procedures and manufacturer's instructions.
Cell Counts.
InvloxP and Prx1-Cre mice strains were combined with the reporter strain Z/EG to obtain invloxP/+; Prx1-Cre; Z/EG and Prx1-Cre; Z/EG embryos. Forelimbs buds were dissected and separately submitted to collagenase treatment (500 U/ml, 60 min at 37°C) to dissociate cells. GFP positive cells were counted using a Bright-Line hematocytometer (Reichert) observed with a GFP filter on a Leica DM6000B.
Chromosome Quantification.
The integrin α6 (Itgα6) locus was used as a marker to quantify each chromosome 2 by real-time PCR. The wild-type Itgα6 allele was located in cis to the inverted loxPs and a mutated integrin α6 allele (Itgα6-) (11) was on the other chromosome. Real-time PCR was performed using TaqMan probes and primers specific for mutated and wild-type Itgα6. DNA was purified from six forelimb buds isolated from, respectively, invloxP/Itgα6- and invloxP/Itgα6-; Prx1-Cre embryos at e10.5. Taqman real-time PCR was carried out according to the manufacturer's protocol (Applied Biosystems). Hoxa13 (Chr6) quantification was used as reference for normalization. See supporting information (SI) Text for primers and probes sequences.
Fluorescent in Situ Hybridization.
Cells from dissected e10.5 forelimb buds were dissociated using collagenase, and treated following classical procedures to obtain interphasic and mitotic chromosome preparations (21). In situ hybridization was performed following standard protocol (22). BACs used as template for probe synthesis were RP24–63014 and RP23–463J10 for chromosome 2 (used separately in independent experiments), and RP23–125F3 for chromosome 19. Biotin and digoxigenin probes were generated by nick translation (Roche), following the manufacturer's instructions, and detected with streptavidin-alexa 546 (Molecular Probes) and anti-DIG antibody (Roche), respectively. Analysis of Chr2-specific hybridization signals was restricted to cells that accurately hybridized with Chr19-specific probe. Two hybridization dots in close vicinity to each other, likely corresponding to replicated loci, were scored as one signal.
Detection and Quantification of DNA Inversion.
InvloxP/+ and invloxP/+; LMOP-Cre eyes were collected from P9 and P22 animals. Genomic DNA was extracted, then purified using QIAquick Kit (Qiagen). We determined DNA concentration accurately using Nanodrop 1000 (Thermo Scientific‘) and diluted DNA to 10 ng/μL in Tris 10 mM pH8.0, 1 μg/μL RNaseA. Quantitative real-time PCR analyses were carried out with Quantitect SYBR Green PCR Kit (Qiagen) on a Mx3000P cycler (Stratagene) following the manufacturer's instructions. Standard curves for quantification were generated from dilution series of genomic DNA or purified “BD” PCR fragment. Primers pair encompassing wild-type Hoxd13 locus was used as a reference for normalization. Photoreceptors correspond to 72% of the total cell population of the retina (23). Therefore raw data were therefore divided by 0.72 to obtain the actual percentage of inverted allele within the photoreceptor cell population (see SI Text data for primer sequences).
Supplementary Material
Acknowledgments.
We thank Annie Dumouchel, Ira Roshan-Afshar, and Marie-France Rousseau for technical assistance; members of the laboratory for sharing reagents; and Michel Cayouette, Jacqueline Deschamps, Denis Duboule, and Mark Featherstone for comments and suggestions. We also thank Rolf Zeller and Artur Kania for critically reading the manuscript. We are very grateful to Mylène Docquier for advice on real-time PCR. BACs for FISH were obtained from Federico Gonzalez and Aurelio Balsalobre. This work was supported by the Canadian Institutes for Health Research (CIHR-82880), a Canada Research Chair (to M.K.), and by a postdoctoral fellowship from the Fondation pour la Recherche Médicale (to D.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807484105/DCSupplemental.
References
- 1.Yu Y, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet. 2001;2:780–790. doi: 10.1038/35093564. [DOI] [PubMed] [Google Scholar]
- 2.Herault Y, Rassoulzadegan M, Cuzin F, Duboule D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE) Nat Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- 3.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 4.Lewandoski M, Martin GR. Cre-mediated chromosome loss in mice. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- 5.Matsumura H, et al. Targeted chromosome elimination from ES-somatic hybrid cells. Nat Methods. 2007;4:23–25. doi: 10.1038/nmeth973. [DOI] [PubMed] [Google Scholar]
- 6.Magnuson T, et al. The early lethality of autosomal monosomy in the mouse. J Exp Zool. 1985;236:353–360. doi: 10.1002/jez.1402360313. [DOI] [PubMed] [Google Scholar]
- 7.Kmita M, Kondo T, Duboule D. Targeted inversion of a polar silencer within the HoxD complex re-allocates domains of enhancer sharing. Nat Genet. 2000;26:451–454. doi: 10.1038/82593. [DOI] [PubMed] [Google Scholar]
- 8.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 9.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 10.Kmita M, Fraudeau N, Herault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420:145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- 11.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 12.Gimond C, et al. Cre-loxP-mediated inactivation of the alpha6A integrin splice variant in vivo: Evidence for a specific functional role of alpha6A in lymphocyte migration but not in heart development. J Cell Biol. 1998;143:253–266. doi: 10.1083/jcb.143.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le YZ, et al. Mouse opsin promoter-directed Cre recombinase expression in transgenic mice. Mol Vis. 2006;12:389–398. [PubMed] [Google Scholar]
- 14.Palmiter RD, et al. Cell lineage ablation in transgenic mice by cell-specific expression of a toxin gene. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 15.Breitman ML, et al. Genetic ablation: Targeted expression of a toxin gene causes microphthalmia in transgenic mice. Science. 1987;238:1563–1565. doi: 10.1126/science.3685993. [DOI] [PubMed] [Google Scholar]
- 16.Ivanova A, et al. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis. 2005;43:129–135. doi: 10.1002/gene.20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito M, et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- 18.Buch T, et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 19.Dor Y, Melton DA. How important are adult stem cells for tissue maintenance? Cell Cycle. 2004;3:1104–1106. [PubMed] [Google Scholar]
- 20.Furukawa N, Saito M, Hakoshima T, Kohno K. A diphtheria toxin receptor deficient in epidermal growth factor-like biological activity. J Biochem. 2006;140:831–841. doi: 10.1093/jb/mvj216. [DOI] [PubMed] [Google Scholar]
- 21.Akeson E, Davisson M. Mitotic chromosome preparations from mouse cells for karyotyping. Curr Protocols Hum Genet. 2000:1–19. doi: 10.1002/0471142905.hg0410s25. 4.10. [DOI] [PubMed] [Google Scholar]
- 22.Bayani J, Squire J. Fluorescence in situ hybridization (FISH) Curr Protocols in Cell Biol. 2004:1–52. doi: 10.1002/0471143030.cb2204s23. 22.4. [DOI] [PubMed] [Google Scholar]
- 23.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.