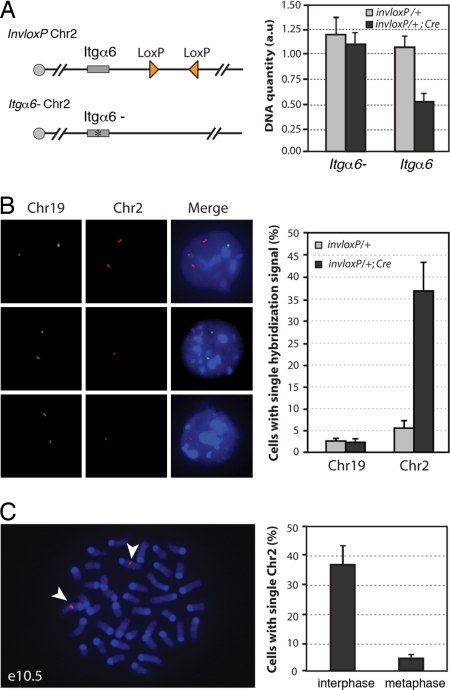
Fig. 2.
TRIP results in chromosome loss and cell death in proliferating cells. (A) Real-time PCR detection of loss of the loxP-carrying Chr2. Chr 2 carrying inverted loxP sites (triangles) was combined with a Chr2 with a mutation in the integrin α6 gene (Itgα6-) (see Materials and Methods). Integrin α6 quantification values were normalized using the Hoxa13 gene located on Chr6. Data are presented as means and ranges from two independent experiments. (B) FISH detection of Chr2 monosomy. In e10.5 invloxP/+; Prx1-Cre limb buds, some cells showed two hybridization signals for Chr2 (Left, top row) while others had a single hybridization signal (Left, middle and bottom rows). For each experiment, at least 200 cells for each genotype were analyzed. Data are presented as means ± SD from three independent experiments. Background level of cells with single signal for Chr19 or Chr2 (3–6%) most likely corresponds to technical limitation or FISH signals in close vicinity to each other (see Materials and Methods) (C) FISH detection of Chr2 in metaphase cells from invloxP/+; Prx1-Cre forelimb buds at e10.5. More than 50 metaphases were analyzed. Representative metaphase with two copies of Chr2 (arrowheads). Data are presented as means ± SD from three independent experiments. Green, DIG probe; red, biotin probe.