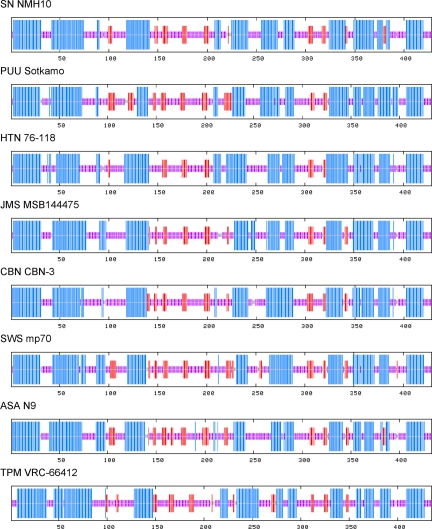
Fig. 2.
Consensus secondary structure of N protein of ASAV and representative rodent- and soricid-borne hantaviruses, predicted using a high-performance method implemented on the NPS@ structure server (47). As shown, the ASAV N protein was very similar to that of other hantaviruses, characterized by the same coiled-coil helix at the amino terminal end and similar secondary structure motifs at their carboxyl terminals. The predicted structures were represented by colored bars to visualize the schematic architecture: α-helix, blue; β-sheet, red; coil, magenta; unclassified, gray. For simplicity, turns and other less frequently occurring secondary structural elements were omitted. All sequences are numbered from Met-1.