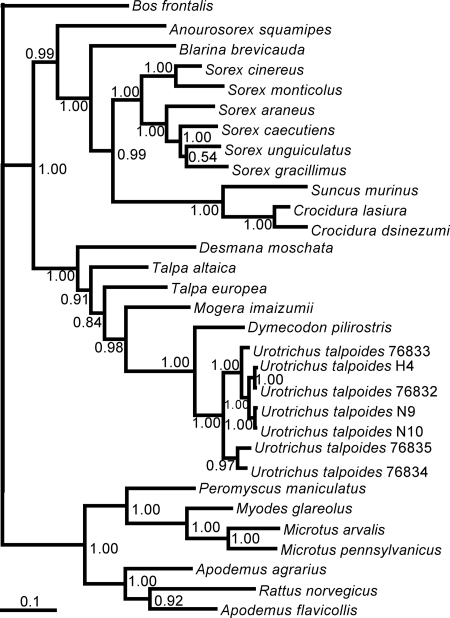
Fig. 4.
Confirmation of host identification of ASAV-infected Urotrichus talpoides by mtDNA sequencing. Phylogenetic tree, based on the 1,140-nucleotide cytochrome b (cyt b) gene, was generated by the ML method. The phylogenetic positions of Urotrichus talpoides H4 (EU918369), N9 (EU918370), and N10 (EU918371) are shown in relationship to other Urotrichus talpoides cyt b sequences from GenBank (Ut76835: AB076835; Ut76834: AB076834; Ut76833: AB076833; Ut76832: AB076832), as well as other talpids, including Desmana moschata (AB076836), Talpa altaica (AB037602), Talpa europea (AB076829), Mogera imaizumii (AB037616), Dymecodon pilirostris (AB076830), and Bos frontalis (EF061237). Also shown are representative murinae rodents, including Apodemus agrarius (AB303226), Apodemus flavicollis (AB032853), and Rattus norvegicus (DQ439844); arvicolinae rodents, including Microtus arvalis (EU439459), Myodes glareolus (DQ090761), and Microtus pennsylvanicus (AF119279); and a neotominae rodent, Peromyscus maniculatus (AF119261), as well as crocidurinae shrews, including Suncus murinus (DQ630386), Crocidura lasiura (AB077071), and Crocidura dsinezumi (AB076837); and soricinae shrews, including Anourosorex squamipes (AB175091), Blarina brevicauda (DQ630416), Sorex cinereus (EU088305), Sorex monticolus (AB100273), Sorex araneus (DQ417719), Sorex caecutiens (AB028563), Sorex unguiculatus (AB028525), and Sorex gracillimus (AB175131). The numbers at each node are posterior node probabilities based on 30,000 trees: two replicate MCMC runs consisting of six chains of 3 million generations each sampled every 1,000 generations with a burn-in of 7,500 (25%). The scale bar indicates nucleotide substitutions per site.