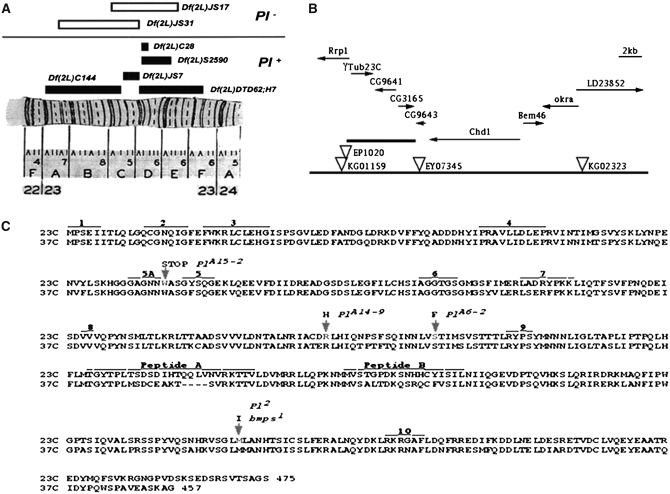
Figure 2.—
Genetic and molecular characterization of the Pearl region in 23C. (A) Deficiency mapping of the Pearl region. (B) Genomic map of region 23C. Predicted transcriptional units in the region are shown by arrows indicating their transcriptional direction. P-element transposons in the region are indicated by inverted triangles. EP1020 is P{EP}Rrp11020, KG01159 is P{SUPor-P}Rrp1KG01159, and EY07345 is P{EPgy2}CG9643EY07345. The thickest line marks the 7-kbp region where γTub23CPl-2 was localized by P-induced male recombination. (C) The amino acid sequences for the two γ-tubulin proteins (labeled 23C and 37C) in Drosophila (Wilson et al. 1997) and the changes found in the γTub23C mutant alleles. The arrows indicate the changes in the γTub23C mutant alleles. At the top of each arrow are the allele name and the amino acid substitution. The numbered areas indicate peptides with known or presumed functions (taken from Burns 1995 and references therein): 1 is a peptide implicated in the autoregulation of β-tubulin translation; peptides 2–10 are implicated in the binding or the hydrolysis of GTP by β-tubulin. Peptide 10 is also implicated in the release of α-, β-, and γ-tubulin from the TCP1α chaperonine. Note that none of the γTub23C mutations are in any of the peptides with known or presumed functions and that all the changed residues in the γTub23C mutants are identical in both γTub23C and γTub37C wild-type genes.