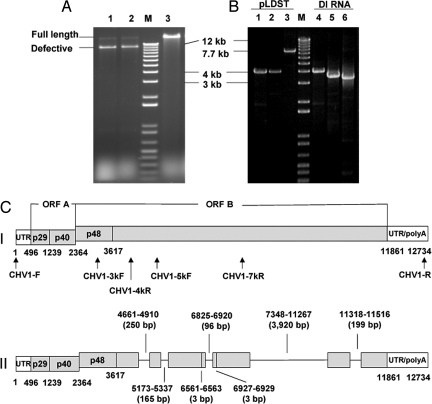
Fig. 1.
Accumulation and characterization of hypovirus DI RNAs. (A) Agarose gel (1%) banding pattern of hypovirus dsRNAs isolated from hypovirus CHV1-EP713-infected wild-type C. parasitica strain EP155 (lane 1, 10 subcultures), dicer 1 gene deletion mutant strain Δdcl-1 strain (lane 2, 10 subcultures), and dicer-2 gene deletion mutant strain Δdcl-2 (lane 3, 17 subcultures). The migration positions of full-length CHV1-EP713 dsRNA and ≈8-kb DI RNA, formerly designated M-RNA (11), are shown in the left margin. The lane marked “M” contains the 1-kb ladder DNA size markers (Gibco). (B) PCR-amplified fragments from full-length CHV1-EP713 cDNA template plasmid pLDST (lanes 1–3) and from cDNA template generated from isolated ≈8 kb CHV1-EP713 DI RNA (lanes 4–6). Primer pairs CHV1-F/CHV1–4kR, CHV1–3kF/CHV1–7kR, and CHV1–5kF/CHV1-R were used, respectively, to amplify regions of the CHV1-EP713 genome corresponding to the 5′ terminus to 4 kb (lanes 1 and 4), 3 kb–7 kb (lanes 2 and 5), and 5 kb–12.7 kb (the 3′ terminus; lanes 3 and 6). The rational for primer selection was as follows. Primers spaced 1 kb along the viral genome RNA were tested for amplification of cDNA generated from isolated DI RNA. This exercise gave an estimate of the location of deleted regions. A subset of these primers (above) predicted to produce a set of overlapping PCR products that spanned the DI RNA was then selected. (C) Diagram of the structure of the major 8,098-nt-long CHV1-EP713-derived DI RNA shown in Fig. 1A, relative to the full-length parental viral RNA. The genetic organization of the parental hypovirus CHV1-EP713 RNA is shown at the top (I). The coding strand is 12,712 nucleotides, excluding the poly (A) tract, and contains two ORFs, ORFs A and B (35). ORF A encodes two polypeptides, p29 and p40, that are released from polyprotein p69 by an autocatalytic event mediated by the papain-like protease domain with p29 (36). An autocatalytic event also releases p48, a second papain-like protease, from the N terminus of the ORF B polyprotein, which contains RdRp and helicase domains. The positions of primer pairs used to amplify fragments shown in B are indicated below the full-length CHV1-EP713 RNA diagram. In (II), the positions of seven deletions found in the 8,098-nt DI RNA are indicated as lines, whereas the regions retained in the DI RNA are indicated by gray bars. The coordinates and size of each deletion is indicated above or below the DI RNA diagram.