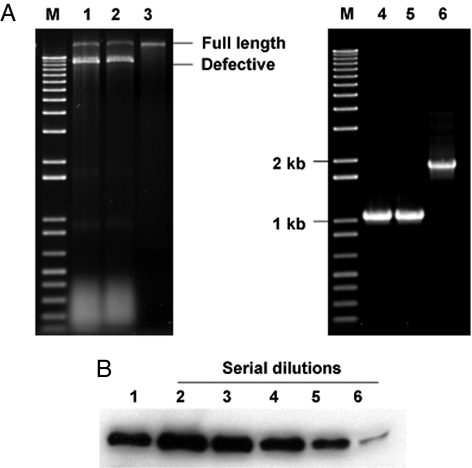
Fig. 3.
Analysis of recombinant hypvoirus Ctp40[2AEGFP] stability and directed EGFP expression levels after 10 subcultures. (A Left) Agarose (1%) gel analysis of dsRNAs recovered from Ctp40[2AEGFP]-infected wild-type strain EP155 (lane 1), dicer-1 mutant strain Δdcl-1 (lane 2), and dicer-2 mutant strain Δdcl-2 (lane 3). The lane marked “M” contains 1-kb DNA size markers. The migration positions of full-length Ctp40[2AEGFP] RNA and DI RNAs are indicated on the right. (Right) RT-PCR analysis of the Ctp40[2AEGFP] RNA recovered from wild-type strain EP155 (lane 4) and mutant strains Δdcl-1 (lane 5) and Δdcl-2 (lane 6) for the presence of the 2A/EGFP nonviral sequences. The region containing the 2A/EGFP sequences in the original recombinant Ctp40[2AEGFP] RNA used to initiate infections was amplified with primer pairs CHV1–2kF and CHV1–3kR corresponding to the CHV1-EP713 map positions 2 kb and 3 kb that flank the insertion site. (B) Endpoint dilution immunoblot analysis of EGFP expression directed by vector Ctp40[2AEGFP] in strain Δdcl-2 relative to that produced in the EGFP-transformed strain EP155/EGFP-Bn, similar to that reported by Parsley et al. (18). Lane 1, 1 μg of total protein from EP155/EGFP-Bn; lanes 2–6, 1 μg, 400 ng, 200 ng, 100 ng, and 50 ng of protein, respectively, from the Δdcl-2/Ctp40[2AEGFP] strain (1-, 2.5-, 5-, 10-, and 20-fold dilution, respectively). The EGFP expressed in the transgenic EP155/EGFP-Bn strain and the Ctp40[2AEGFP]-infected strain migrated with the same mobility and consistent with a size of 27 kDa.