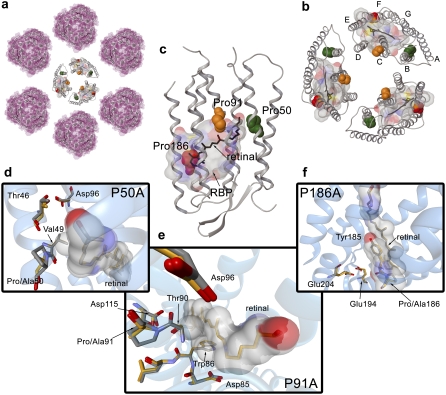
FIGURE 1.
Halobacterium salinarum PM. (a) Two-dimensional paracrystalline arrangement of the PM disposed in hexamers. (b) Trimer of bR molecules (PDB code 1brr). The chromophore molecule of retinal is shown in black. Helix-embedded prolines are indicated by Van der Waals spheres in green (Pro50), orange (Pro91), and red (Pro186). To identify the RBP, a molecular surface around the retinal with a 5 Å radius has been calculated. The color code for the molecular surface corresponds to the atom type (carbon, gray; nitrogen, blue; oxygen, red; sulfur, yellow). (c) Representation of the 3D structure of bR indicating the presence of the seven transmembrane α-helices and the β-strands in a cartoon visualization. The figure has been generated from the 1c3w PDB code following the same color code as for (b). (d) Overlay of the chain A of 1py6 and 1pxr crystal structures obtained at pH 3.5. (e) Overlay of the chain A of 1py6 and 1q5j crystal structures obtained at pH 3.5 and 3.7, respectively. (f) Overlay of the chain A of 1py6 and 1q5i crystal structures obtained at pH 3.5 and 4.0, respectively. For distinction, the carbon atoms in 1py6 (WT) are plotted in gray and carbon atoms in 1pxr (P50A), 1q5j (P91A), and 1q5i (P186A) are plotted in orange. Retinal and residues interacting with the chromophore are indicated in addition by a molecular surface. Molecular representations in (a), (b), and (c) have been plotted with the academic version of MAESTRO 8.0 software. Pictures in (d), (e), and (f) have been generated with BallView.