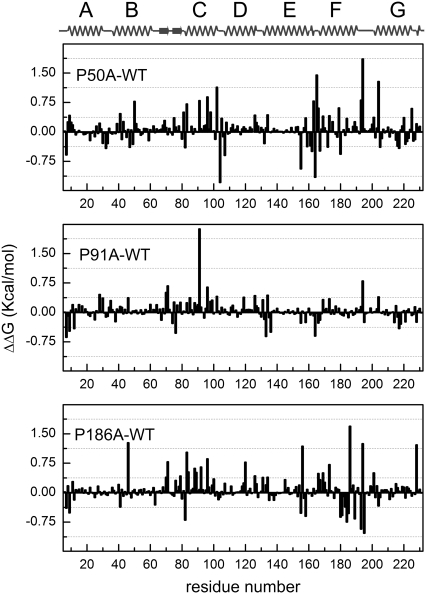
FIGURE 2.
FoldX residual stability of helix-embedded prolines. Calculated residual stabilities from the available crystal structures (average of the chains A and B) for WT (PDB code 1py6), P50A (PDB code 1pxr), P91A (PDB code 1q5j), and P186A (PDB code 1q5i). To determine the changes in the WT bR stability, each residual stability value of the mutation was subtracted to the corresponding value in the structure of the native protein. Values beyond ±0.46 kcal/mol23 indicate significant stabilization (negative values) or destabilization (positive values) induced by the mutation. A linearization of the 3D structure is shown on the top of the figure for a clearer visualization. For the overall structure stabilities calculated with FoldX for the different mutations, refer to Table 1.