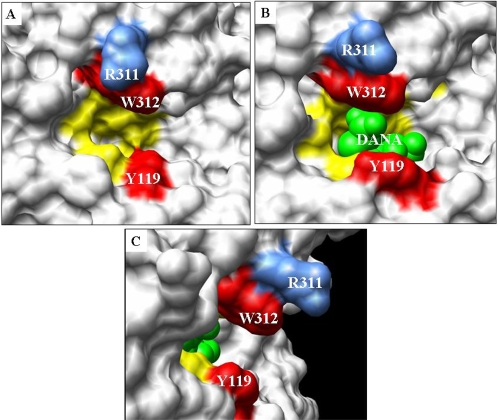
Figure 5. The epitope recognized by inhibitory llama antibodies maps to the TcTS catalytic site.
View of the TcTS active site shown as surface representation using the program Chimera. (A) Free TcTS, (B) DANA-TcTS (shows the conformational change upon binding of DANA to TcTS) and (C) shows a 90° rotation of TcTS-DANA structure highlighting the arginine 311 residue protruding from the active site. PDBs used are 1MS4 and 1MS8. Residues involved in the catalytic site are colored as follows: mutated residues that were analyzed in this work (Trp312 and Tyr119) in red, other catalytic amino acids (Arg35, Asp59, Asp96, Met98, Arg314, Arg245, Glu230 and Tyr342) in yellow, space-fill model of DANA in green and Arg311 in blue.