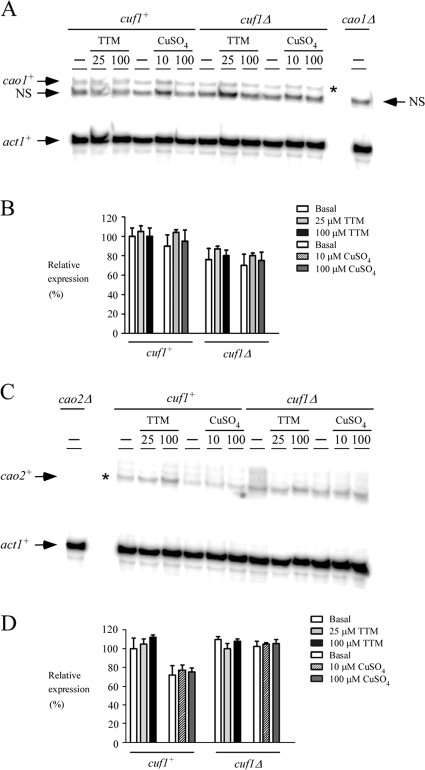
FIG. 2.
cao1+ and cao2+ mRNA levels are constitutively expressed and present in cuf1Δ cells. (A) The indicated isogenic strains were grown to logarithmic phase in yeast extract plus supplements. Cultures were incubated in the absence (−) or presence of TTM (25 and 100 μM) or CuSO4 (10 and 100 μM) for 1 h at 30°C. Total RNA was isolated and analyzed by RNase protection assays. Steady-state mRNA levels of cao1+ and act1+ (indicated with arrows) were analyzed in strains expressing (cuf1+) or lacking (cuf1Δ) the cuf1+ allele. As a control, the cao1+ mRNA was not detected in the isogenic cao1Δ null strain. NS, nonspecific signal. The asterisk indicates the cao1+ mRNA. (B) Quantification of results of three independent RNase protection assays, including results shown in panel A. (C) Aliquots of the cultures described for panel A were examined by RNase protection assays for steady-state levels of cao2+ mRNA. The arrows indicate signals corresponding to cao2+ and act1+ steady-state mRNA levels. RNA isolated and analyzed from the isogenic cao2Δ null strain was used as a control. The asterisk indicates the cao2+ mRNA. (D) Quantification of cao2+ mRNA levels after the treatments. The values are the averages from triplicate determinations ± standard deviations.