Figure 1.
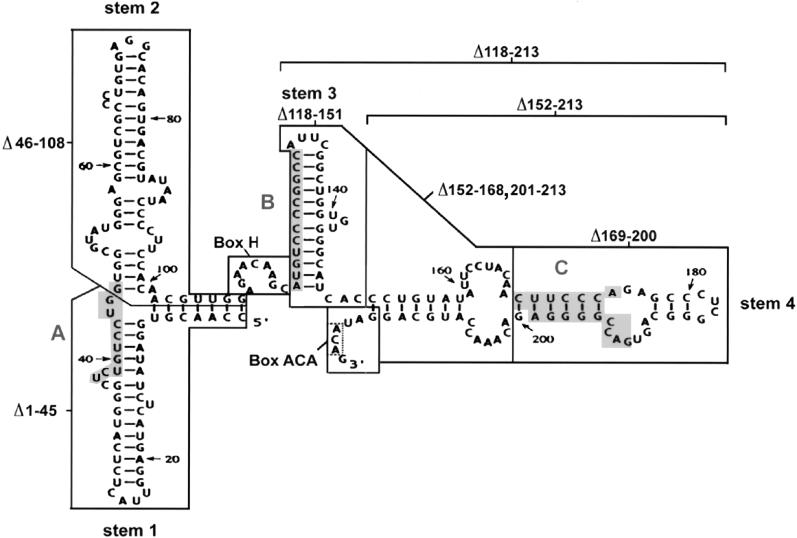
Structure and mutations of U17 snoRNA. The sequence and structure of Xenopus laevis U17 snoRNA (copy f), found in the sixth intron of the gene for ribosomal protein S7 (formerly S8), is from Cecconi et al. (1994), with sequence corrections at positions 33 (G instead of A), 91 and 161 (additional U). Shaded areas A and B in stems 1 and 3 of U17 are complementary to sequences in 18S rRNA (Cecconi et al., 1994). Shaded area C in stem 4 is complementary to the external transcribed spacer of pre-rRNA (Cecconi et al., 1994). The regions of U17 that were mutated in the present study are enclosed by lines. Box ACA, consisting of three nucleotides (enclosed by a dotted line), lies within the single-stranded 3′ end of the molecule. The sequences of the mutants of U17 designed for the present study are listed in the lower portion; nucleotides that are the same as in wild-type U14 snoRNA are shown by dots in the sequence alignment, and deletions are indicated by dashes. The double mutants Δbox H/Δbox ACA+, Δbox H/Δbox ACA, sub. box H/sub. box ACA+, Δ46–108/Δbox H, and Δ46–108/Δ152–213 are not shown, because they are simply the sum of the individual single mutations.