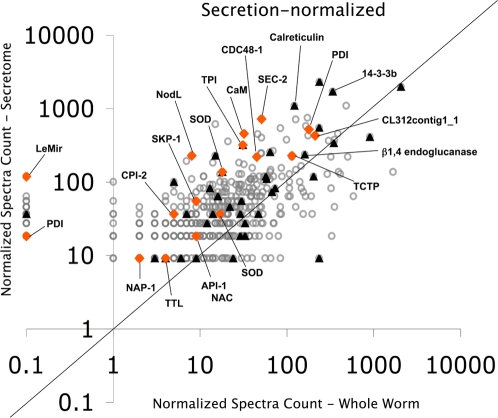
Figure 3. Relative abundance of secreted proteins compared to extracts from intact nematodes.
Spectrum counting was used for relative protein quantification compating the secretome to the intact nematode proteome. The number of valid MS/MS spectra from each protein was normalized to the total MS/MS spectra number of each dataset. The normalized spectrum count ratio of each protein (secretome/whole worm proteome) was used to evaluate if the protein was enriched in the secretome. Circle represents all secreted proteins identified in this study; Triangle represents proteins previously reported to be in the secretome (A complete description of these proteins is shown in Table S5); Diamond represents proteins that we found in M. incognita secretome and that could play a crucial role in the establishment of the host-pathogen compatibility. These proteins are discussed in detail.