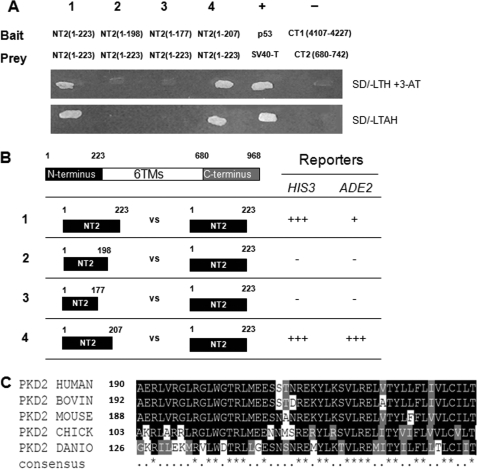
FIGURE 3.
Dimerization of the polycystin-2 N terminus (NT2) detected by yeast two-hybrid assays. A, growth of yeast co-transformants cultured on selective media S.D./-LTH with 2 mm 3-AT and S.D./-LTAH to activate HIS3 and ADE2 selection markers, respectively. The pairs of NT2 sequences tested are numbered from 1–4 and their sequences indicated. Constructs pGBKT7–53 (p53) and pGBADT7-T (SV40-T antigen) were used as a pair of positive controls while pGBAD-B-CT1 (4107–4227) and pACT2-B-CT2 (680–742) were used as a pair of negative controls. B, truncations of the N terminus of polycystin-2 (NT2) marked by the number of starting and ending amino acid residue and their interaction with NT2. The fragments on the left column are in bait (BD) constructs, while the fragments in the right column are in prey (AD) constructs. The numbers in the left column indicate the pairs of NT2 constructs tested and correspond to those shown in A. Positive (interaction) and negative results are indicated by +++, ++, +, and - for the appearance of positive growth on selective medium within 24 h, 48 h, and 3–7 days or no positive growth in 2 weeks, respectively. C, multiple sequence alignment of PC2 orthologues from different species overlapping with the sequence of human NT2-(190–238). Amino acids which show perfect conservation down to zebrafish are indicated by an asterisk (*) in the bottom line. In the region between amino acids 190 and 223, 23 of 33 amino acids (70%) are identical or similar. This contrasts with the region between amino acids 119 and 189 where only 1 of 70 amino acids (1.4%) show identity from human to Danio. The accession numbers for each sequence are as follows: Q13563 (Homo sapiens), O35245 (Mus musculus), Q4GZT3 (Bos taurus), Q5ZM00 (Gallus gallus), and Q6IVV8 (Danio rerio).