FIGURE 3.
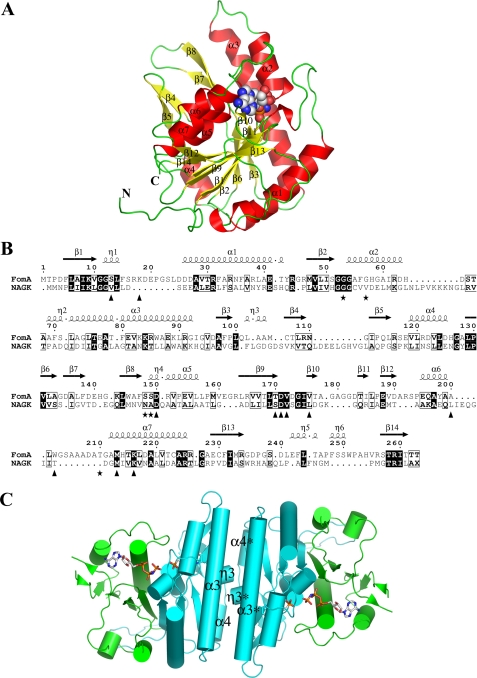
The structure of the FomA·MgAMPPNP·fosfomycin complex. A, ribbon diagram of a monomer of FomA withα-helices andβ-strands labeled. The AMPPNP molecule is shown as a space filled model. B, structure based sequence alignment between FomA and NAGK. The secondary structure elements of FomA defined by DSSP (47) are given on top. Triangles indicate amino acids that line the AMPPNP-binding site, whereas stars indicate fosfomycin-binding site residues. The image was generated using the program ESPript (48). C, cartoon representation of the dimer with the dyadic axis perpendicular to the paper. The N- (cyan) and the C-lobe (green) domains are shown. AMPPNP and fosfomycin substrate molecules are shown in stick representation.