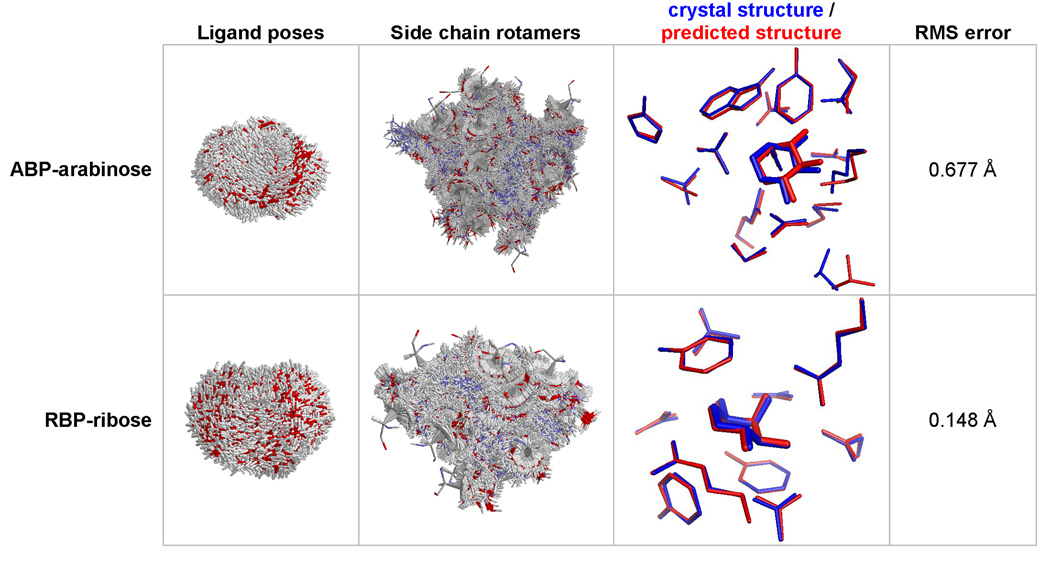
Figure 3.
Prediction of binding site coordinates. Starting from crystal structures stripped of the ligand and the contacting residues, the active site was reconstructed by finding the lowest energy arrangement of the ligand and side chains. For ABP-arabinose (PBD code: 6ABP), the coordinates of the arabinose and 15 contacting residues (10, 14, 16, 17, 64, 89, 90, 108, 145, 147, 151, 204, 205, 232, 259) were predicted using 6028 rotamers per position and 4111 ligand poses. For RBP-ribose (PDB code: 2DRI), the coordinates of ribose and 10 contacting residues (13, 15, 16, 89, 90, 141, 164, 190, 215, 235) were predicted using 5449 rotamers per position, and 4639 ligand poses.