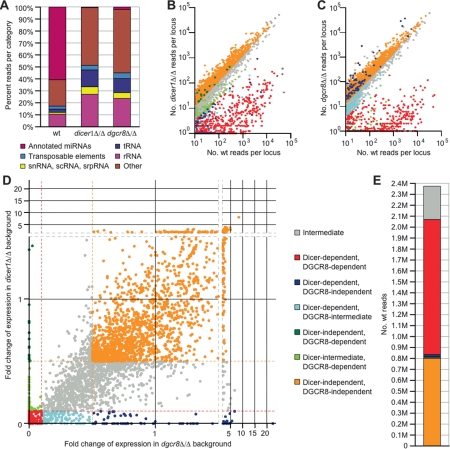
Figure 1.
DGCR8 and Dicer dependence of mouse small RNAs. (A) Classification of small RNAs based on UCSC mouse genome annotations (mm8). See Supplemental Table 1 for a tabular representation of the data. (B) Read counts from the wild-type versus dicer1Δ/Δ libraries for genomic loci with at least 10 reads from the wild-type library, colored as in D. (C) Read counts from the wild-type versus dgcr8Δ/Δ libraries for genomic loci defined in B, colored as in D. (D) Dependencies of the small RNAs derived from genomic loci defined in B. Each axis indicates the following quotient: read count from mutant library/read count from wild-type library, with read counts normalized to the number of t/sn/sc/srpRNA-derived reads from that library. Ratios <0.1 defined enzyme dependence (red hashed line) and ratios >0.5 defined enzyme independence (gold hashed line). Loci were categorized in consideration of their dependencies on both Dicer and DGCR8, and are color coded according to the key. (E) The number of reads from each dependency category from D in the wild-type library. See Supplemental Table 3 for tabular representation.