Figure 3.
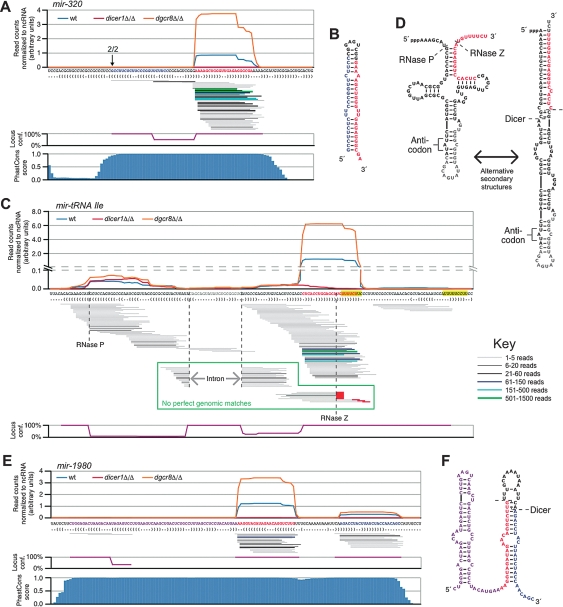
Small RNAs that are Dicer-dependent and DGCR8-independent and derive from highly conserved shRNA. (A) Small RNAs from mir-320 were DGCR8-independent, Dicer-dependent. (Top) The normalized read count from each library, plotted above the genomic sequence and predicted secondary structure, as in Figure 2A. RNA species are represented below as lines, with thickness/color representing the number of reads (see the key). Below that is the mean locus confidence for small RNA reads at each position; 100% represents a unique match to the genome; percentages are then divided by the mean number of genomic loci to which the sequences map. (Bottom) PhastCons score at each nucleotide position. The arrow represents the 5′ end as determined by RACE in dicer1Δ/Δ and the number above represents the number of clones observed. (B) Predicted secondary structure of the mir-320 pre-miRNA. (C) Small RNAs mapping to a tRNA-Ile gene, shown as in A. Green boxed area represents reads that do not match the genome due to tRNA processing, and red lines represent untemplated CCA addition. mir-tRNA Ile has been annotated as mir-1983. (D) Manually predicted secondary structures for the tRNA cloverleaf structure and alternative shRNA fold. (E) Small RNAs mapping to mir-1980, presented as in A. (F) Predicted secondary structure of the presumed mir-1980 precursor.