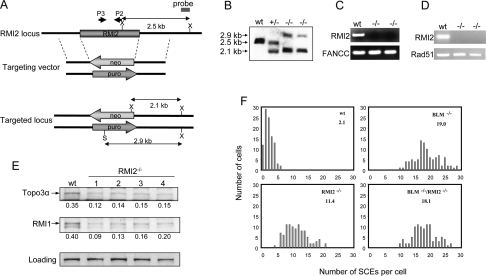
Figure 6.
RMI2-null DT40 cells resemble BLM mutant cells in displaying an elevated level of SCE. (A) Schematic representation of the chicken RMI2 genomic DNA, the two targeting vectors, and the restriction map of the locus after knockout. Two restriction enzyme digestion sites used in Southern Blotting, XhoI and SalI, are marked as X and S, respectively. The primers used for genomic-PCR analysis (P3–P2) are depicted in B. Southern blotting shows that both alleles of the RMI2 gene have been inactivated by the targeting vectors described in A. The genomic DNA from wild-type (wt), RMI2+/−, and RMI2−/− cells was first digested with XhoI and SalI restriction enzymes, and subsequently hybridized with the probe shown in A. (C) Genomic-PCR analysis to show the absence of RMI2 genomic DNA in RMI2−/− cells. The FANCC gene was included as a positive control. (D) RT–PCR analysis to show that RMI2 mRNA is undetectable in RMI2−/− cells. RAD51 was included as a positive control. (E) Immunoblotting to show that Topo 3α and RMI1 levels are reduced in four different clones of RMI2−/− cells. (F) Histograms showing the SCE levels of wild-type, RMI2−/− cells, BLM−/−, and RMI2−/−/BLM−/− cells. The mean number of SCEs per cell is shown in the top right corner.