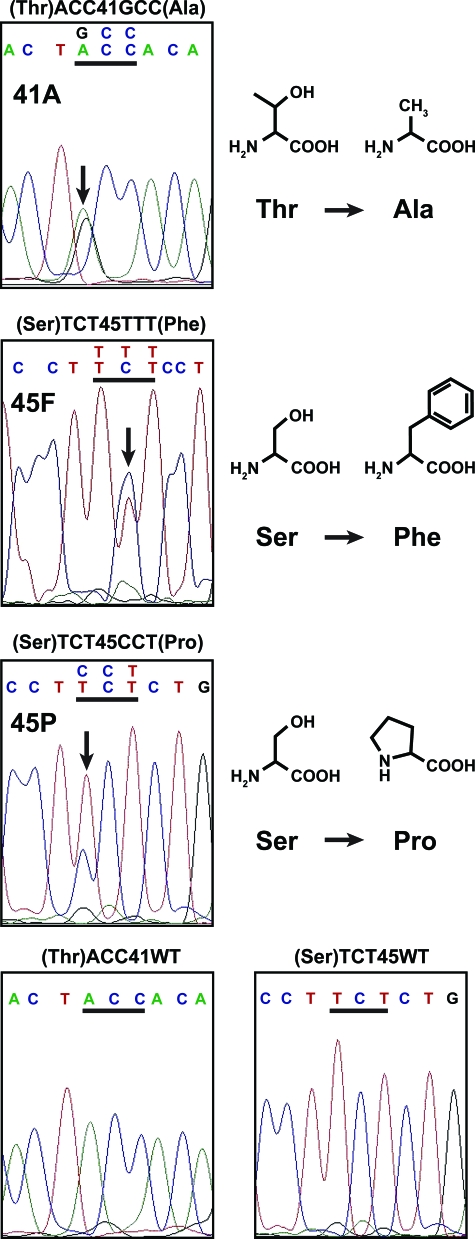
Figure 1.
CTNNB1 exon 3 sequencing of desmoid tumor was available from 154 of the 160 patients (96%) on the TMA. Of these, 138 had sufficient clinical data (and no evidence of FAP) for inclusion and 85% showed one of three specific mutations in CTNNB1. Examples of Sanger sequencing for each mutational type encountered (abbreviated 41A, 45F, and 45P, respectively) and lack of mutation are displayed. In the top three panels, the mutated bp is denoted by a black arrow on the DNA sequence tracing and the codon change is shown directly above the affected codon (underlined in black). In the bottom two panels, unaltered codons 41 and 45 are displayed. Additional data are presented in Table 1. The chemical structures of the amino acids in the normal and mutated codons are depicted on the right. A, adenine; C, cytosine; G, guanine; T, thymine; Thr, threonine; Ala, alanine; Ser, serine; Phe, phenylalanine; Pro, proline; WT, wild type.