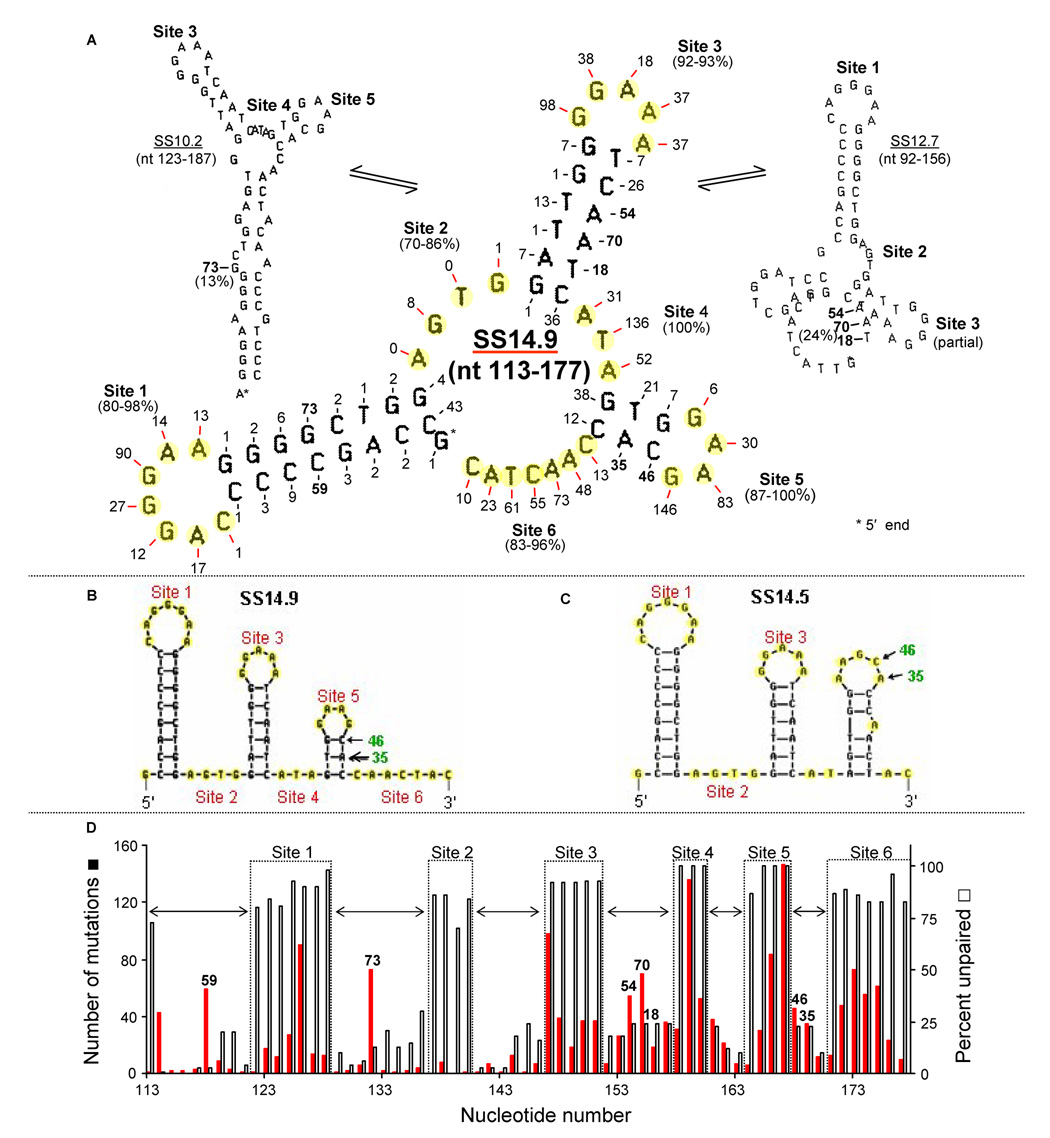
Fig. 2. Inter-conversion of SS14.9 with less stable SSs during transcription.
(A) The dominant SS14.9 flanked by two lower stability structures, SS10.2 (left) and SS12.7 (right). The number of mutations at each unpaired mutable base in SS14.9 is shown in red, and selected mutations in paired bases of stems are in green. SS10.2 is the most stable in which the G in the stem of Site 1 in SS14.9 (73 mutations, in green) and the three bases (AAT, green) in the stem of Site 3 (54, 70, and 18 mutations, respectively, in green), are unpaired. Also shown in green are mutations in the stem of Site 5 (46 and 35) that are reported (mfold) to be in a less stable conformation of nt 113–177. (B) The location of mutations 46 and 35 in SS14.9 depicted by mfold. (C) The less stable SS14.5 configuration of nt 113–177 depicted by mfold. (D) The relationship of mutation frequency and percent unpaired in SS14.9. Horizontal arrows indicate the location of stems, and high mutation numbers (green) in stem sequences indicate mutable bases that are located in SSs other than SS14.9.