Figure 1. 21U-RNAs can be distinguished from other RNA species by their lengths and upstream motif matches.
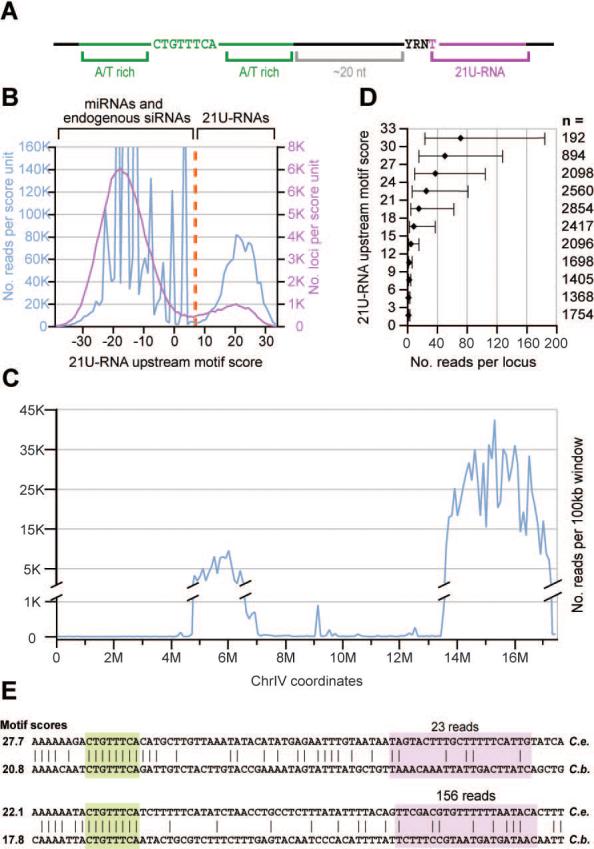
(A) A schematic representation of the 21U-RNA upstream motif as described previously (Ambros et al., 2003; Ruby et al., 2006). (B) The frequency of 21nt RNA reads (blue) or unique sequences (pink) versus upstream motif score. A cut-off score of 7 (orange) was used to define the 21U-RNA population. (C) The distribution of 21U-RNA reads across chromosome IV. Reads were classified as 21U-RNAs by their motif scores and normalized read counts were summed for each non-overlapping 100kb bin (blue). (D) The upstream motif score predicts the magnitude of 21U-RNA expression. For each three-bit bin of motif scores, the number of reads was determined for every observed 21U-RNA locus whose motif score fell in that range. For each bin, the median read number was determined; error bars indicate the 25th and 75th percentiles. The number of loci in each bin is indicated by n. (E) Two 21U-RNA loci whose core upstream motifs are aligned (Blanchette et al., 2004). The core motif (green) and 21U-RNA (pink) are highlighted. The C. briggsae 21U-RNA was annotated based on the highest-scoring 5′ end given the conserved core motif position. The number of reads for the 21U-RNA species from C. elegans is given above, and the motif score for each 21U-RNA ortholog is provided.
