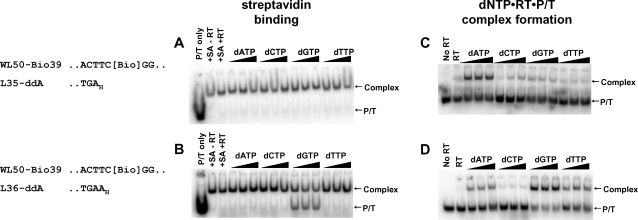
Figure 4. Effect of preformed RT stable ternary complexes on SA binding to a template biotin residue.
Five nM 5′-32P-labeled P/T L35-ddA/WL50-Bio39 (A,C) or L36-ddA/WL50-Bio39 (B,D) were incubated without or with 100 nM RT in the absence of ligands or with 0.8, 3.2 or 6.4 mM dNTP. In A and B, SA (50 nM) was added for 5 min at 37°C, and 0.6 µM biotin was added to bind excess SA. Then RT was dissociated with SDS and urea, and SA-biotin-DNA complexes were separated from free DNA by electrophoresis on nondenaturing gels. For C and D, complexes formed with dNTP were transferred to ice, treated with heparin to dissociate RT•P/T binary complexes and fractionated by nondenaturing gel electrophoresis. Arrows to the right of each panel indicate positions of free DNA (P/T) and SA-biotin-DNA complexes (A,B) or dNTP•RT•P/T ternary complexes (C,D). A portion of each P/T sequence is shown. “Bio” indicates the biotin-ON linkage. Subscript “H” denotes a dideoxyribonucleotide residue.