Abstract
Epigenetic changes provide a frequent mechanism for transcriptional silencing of genes in cancer cells. We previously established that epigenetic mechanisms are important for control of group IIA phospholipase A2 (PLA2G2A) gene transcription in human DU-145 prostate cells. In this study, we analyzed the involvement of such mechanisms in the regulation of five sPLA2 isozymes and the M-type receptor of sPLA2 (sPLA2-R) in human leukemic Jurkat cells. These cells constitutively expressed sPLA2-IB, sPLA2-III, sPLA2-X, and sPLA2-R but not sPLA2-IIA and sPLA2-V. Transcription of sPLA2-IIA and sPLA2-V was, however, detected after exposure of cells to the DNA demethylating agent, 5-aza-2′-deoxycytidine (5-aza-dC). Expression of sPLA2-IIA was further enhanced by additional exposure to interferon-γ and blocked by inhibitors of specificity protein 1, nuclear factor κB, and Janus kinase/signal transducer and activator of transcription-dependent pathways. Sequence analysis and methylation-specific polymerase chain reaction of bisulfite-modified genomic DNA revealed two 5′-CpG sites (-111 and -82) in the sPLA2-IIA proximal promoter that were demethylated after 5-aza-dC treatment. These sites may be involved in the DNA binding of specificity protein 1 and other transcription factors. Similar findings after treatment of human U937 leukemia cells with 5-aza-dC indicate that this mechanism of PLA2G2A gene silencing is not restricted to Jurkat and DU-145 cells. These data establish that regulation of sPLA2-IIA and sPLA2-V in Jurkat and other cells involves epigenetic silencing by DNA hypermethylation.
Introduction
Secreted phospholipases A2 (sPLA2, phosphatide sn-2-acylhydrolases, EC 3.1.1.4) belong to the superfamily of phospholipases that not only catalyze production of bioactive lipid mediators of cell signaling pathways but also function as receptor ligands, independently of any enzymatic activity [1–5]. By way of these functions, the various sPLA2 isozymes play crucial roles in several physiological and pathophysiological processes involving alterations in cellular proliferation, growth, differentiation, and apoptosis. The most thoroughly characterized isozyme, sPLA2-IIA, is constitutively expressed in a variety of cell types including chondrocytes, synoviocytes, mesangial cells, astrocytes, and smooth muscle cells (for review [4]).
Recognition that expression of sPLA2-IIA is increased in numerous cancer cells has formed the basis of the sPLA2-IIA pro-oncogenic activity concept [6–12]. The precise role of the enzyme in tumorigenesis, nevertheless, remains unclear. In human pancreatic cancer cells and gastric adenocarcinoma, expression of sPLA2-IIA is associated with prolonged survival and reduced tendency to metastasis, suggesting antioncogenic properties [13,14]. Moreover, sPLA2-IIA is not appreciably expressed in a number of cancer cell lines including human leukemia Jurkat T cells, monocytic U937 cells [15–17], fibrosarcoma HeLa cells [18], and prostate cancer DU-145 cells [9,19].
Recently, we established that lack of sPLA2-IIA expression and failure of cytokines to induce sPLA2-IIA expression in malignant DU-145 prostate cells can be explained by an epigenetic silencing regulatory mechanism and not by PLA2G2A gene mutations connected with a failed expression of intact sPLA2-IIA protein [19]. Such mechanisms involve aberrant methylation of cytosines belonging to 5′-CpG islands, short stretches of DNA rich in 5′-CpG dinucleotides, and often associated with gene promoters (for review [20,21]). In cancer cells, the methylation may retard expression of genes critical to cell proliferation and apoptosis [21].
With the above considerations in mind, we proposed that epigenetic mechanisms are involved in the silencing of sPLA2-IIA in a number of so-called “sPLA2-IIA-negative” cells. To address this hypothesis, we analyzed the expression of sPLA2-IIA and four other sPLA2 isozymes (IB, III, V, and X) in connection with the M-type receptor of sPLA2 (sPLA2-R) in cytokine-primed Jurkat cell lines before and after treatment with demethylating agent, 5-aza-2′-deoxycytidine (5-aza-dC).We also determined the methylation status of several distinct 5′-CpG sites in the proximal sPLA2-IIA promoter region.
Materials and Methods
Materials
Recombinant human interleukin 1β (IL-1β), interleukin 6 (IL-6), tumor necrosis factor α (TNF-α), and interferon γ (IFN-γ) were purchased from Roche Diagnostics Applied Science (Mannheim, Germany). PD98059, Janus kinase (Jak) inhibitor I, phorbol-12-myristate 13-acetate (PMA), SP-600125, and H-1152 were obtained from Calbiochem (San Diego, CA). Forskolin, 5-aza-dC, mithramycin A, and caffeic acid phenethyl ester (CAPE) were purchased from Sigma-Aldrich (Deisenhofen, Germany). Phorbol-12-myristate 13-acetate, PD98059, forskolin, SP-600125, mithramycin A, Jak inhibitor I, and CAPE were dissolved in dimethyl sulfoxide (DMSO). The final concentrations of solvents were 0.3% or less. Controls using DMSO alone were run in all cases.
Cell Culture and Incubation
Jurkat (human T lymphocyte acute leukemia) and U937 (human hystiocytic lymphoma) cell lines were purchased from German Collection of Microorganisms and Cell Cultures (Berlin, Germany). They were cultured in a standard cell medium RPMI-1640 supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2. For all experiments, exponentially growing subconfluent cells were used at passages 5 to 8.
For 5-aza-dC treatment, cells were cultured in RPMI 1640 cell medium with 10% fetal calf serum and 5-aza-dC added to the final concentration of 1 to 10 µM from a freshly prepared 10-mM stock solution. After 3 days, during which the culture medium containing 5-aza-dC was renewed once each day, the cells were harvested and analyzed for sPLA2-IIA protein and mRNA levels.
RNA Extraction and Reverse Transcription-Polymerase Chain Reaction Analysis of sPLA2 Enzyme Expressions
RNA was isolated after lysis of Jurkat T cells in TRI Reagent (Sigma-Aldrich) according to the manufacturer's instructions. Isolated RNA was converted to cDNA using the GeneAmp RNA-PCR Kit (PerkinElmer LAS GmbH, Jügesheim, Germany). Portions of the reverse-transcribed reaction products were then amplified by polymerase chain reaction (PCR) for the identification of sPLA2-IB, -IIA, -III, -V, -X, the M-type of the sPLA2 receptor (sPLA2-R), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a house-keeping gene. The applied primer pairs are summarized in the Table 1.
Table 1.
Sequences of the Applied Forward (f) and Reverse (r) Primers.
| Genes | Sequences | Size (bp) |
| PLA2G1B | 5′-AAA TGA TCA AGT GCG TGA TCC-3′ (f) | 243 |
| 5′-TTG CTG CTA CAG GTG ATT GC-3′ (r) | ||
| PLA2G2A | 5′-GTG ATC ATG ATC TTT GGC CTA CTG CA-3′ (f) | 411 |
| 5′-TCT CCC TCG TGG GGA GCA ACG ACT-3′ (r) | ||
| PLA2G3 | 5′-ACAACTCTTCTATGCCTGG-3′ (f)′ | 256 |
| 5′-TGTGACATCCCTAACTTCC-3′ (r) | ||
| PLA2G5 | 5′-GGG CTG CAA CAT TCG CAC AC-3′ (f) | 278 |
| 5′-CCT CTC TCA GGA ACC AGG CAG-3′ (r) | ||
| PLA2G10 | 5′-CCA TCG CCT ATA TGA AAT ATG G-3′ (f) | 295 |
| 5′-TAG GAA CTG GGG GTA GAA GAG-3′ (r) | ||
| PLA2G1R | 5′-CAG AAG AAA GGC AGT TCT GGA TTG-3′ (f) | 496 |
| 5′-AAA GCC ACA TCT CTG GCT CTG ATT-3′ (r) | ||
| GAPDH | 5′-CGG AGT CAA CGG ATT TGG TCG TAT TG-3′ (f) | 439 |
| 5′-GCA GGA GGC ATT GCT GAT GAT CTT G-3′ (r) |
The oligonucleotides used for analysis of mRNA were synthesized according to published nucleotide sequences of human sPLA2-IB, sPLA2-IIA, sPLA2-III, sPLA2-V, sPLA2-X, sPLA2-R, and GAPDH genes (http://www.ensembl.org/index.html). Primer pairs for PCR were applied in a final concentration of 0.8 µM. The conditions for amplification were as follows: 40 cycles at 94°C for 30 seconds, 68°C for 30 seconds, and 72°C for 1 minute. Buffers and reagents were from GeneAmp Kit (PerkinElmer LAS GmbH). Amplified products were analyzed by electrophoresis on agarose gels.
Quantitative Determination of sPLA2-IIA by Enzyme-Linked Immunosorbent Assay
Release of sPLA2-IIA protein into the culture medium was determined using an enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's instructions (Cayman Chemical, MI). Culture medium was removed, centrifuged for 10 minutes at 400g to remove cell debris, and used for sPLA2-IIA protein determinations. Total cell protein was determined using a bicinchoninic acid assay kit (Sigma-Aldrich).
Searching for 5′-CpG Islands and Potential Binding Sites for Transcription Factors in Proximal Promoter Regions of sPLA2 Isozymes and M-type Receptor of sPLA2
To address the question of whether 5′-CpG islands are present in the promoters of the sPLA2 isozymes and sPLA2-R, we used MethPrimer software (http://www.urogene.org/methprimer) to analyze the promoter regions -1000 to +200 relative to the transcription starting sites of each gene [22]. We also used TFSEARCH software [23,24] and an object-oriented transcription factors search (ooTFD) software [25] to assess whether the proximal promoter of PLA2G2A contains binding sites for transcription factors known to be crucial for the sPLA2-IIA expression.
DNA Methylation Analysis of Selected 5′-CpG Sites in the Proximal sPLA2-IIA Promoter Region
Genomic DNA was extracted from control and 5-aza-dC-treated Jurkat cells using DNazol (Invitrogen Life Technologies, Karlsruhe, Germany) following the manufacturer's instructions. Sodium bisulfite treatment of the genomic DNA was performed using the Methyl-Detector kit from Active Motif (Carlsbad, CA) [26–28].
To amplify the bisulfite-modified DNA, a seminested PCR was performed with primers covering regions within the proximal promoter region of the sPLA2-IIA gene from -351 to +152 relative to the transcription start site [29]. As extrinsic primers, 5′-CTC ATA CAT ATC AAA TCA T-3′ and 5′-GTA ATT GGT AGT TTT TTT G-3′, and as intrinsic primers, 5′-CTC ATA CAT ATC AAA TCA T-3′ and 5′-GTA AAT GAG TTT ATA GTT TG-3′, were applied in a final concentration of 0.8 µM. The conditions of PCR were as follows: 20 cycles at 95°C for 45 seconds, 44°C for 60 seconds and 72°C for 45 seconds using the extrinsic primers, and 35 cycles at 95°C for 30 seconds, 45°C for 45 seconds, and 72°C for 30 seconds using the intrinsic primers. The buffers and reagents were from GeneAmp Kit (PerkinElmer LAS GmbH). The PCR products were purified using a QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany). DNA sequences of both strands were analyzed with the ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems, Foster City, CA). To verify the efficiency of bisulfite treatment, the sequence of bisulfite-treated genomic DNA was compared with that of untreated genomic DNA. As a further control, Jurkat DNA treated in vitro with Sss I methyltransferase and purchased from New England BioLabs Inc. (Ipswich, MA) was used.
In addition to sequence analysis of bisulfite-modified genomic DNA, methylation-specific PCR was performed. For this purpose, the following forward (f) and reverse (r) primers were designed and applied:
5′-TGG TAT TAG TTA TTG ATA CGT-3′, f for methylated 5′-CpG (-186);
5′-CGG TAT TAG TTA TTG ATA TGT-3′, f for unmethylated 5′-CpG (-186);
5′-ATT ATT TAG GGG TAT GGG CGA-3′, f for methylated 5′-CpG (-111);
5′-ATT ATT TAG GGG TAT GGG TGA-3′, f for unmethylated 5′-CpG (-111);
5′-TTT TGA GTT TAT TAA TTG ATT ACG T-3′, f for methylated 5′-CpG (-82);
5′-TTT TGA GTT TAT TAA TTG ATT ATG T-3′, f for unmethylated 5′-CpG (-82);
5′-CTC CAA AAT TAT ATC CCC AAA-3′, r primer.
The conditions of amplification for methylation analysis were as follows: 40 cycles at 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds in the case of the primer pairs to analyze the 5′-CpG sites at positions -186 and -82. In case of the 5′-CpG site at position -111, a touchdown PCR with an annealing temperature between 45 and 50°C was performed. Finally, PCR products were analyzed by electrophoresis on agarose gels.
Data Analysis
Data were analyzed by one-way analysis of variance coupled with Dunnett's post hoc test to compare each experimental group with a nominated control group using SPSS 14.0 software. Differences were considered significant at P < .05.
Results
Basal and Cytokine-Induced Expression of Secreted Phospholipases A2 and Phospholipase A2 Receptor in Human Jurkat Cells
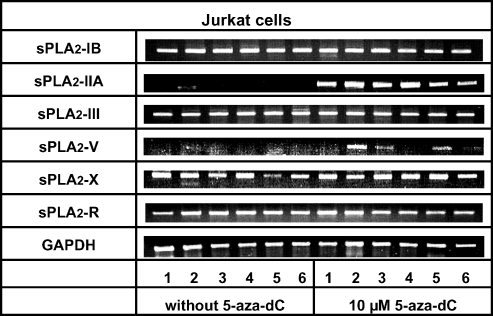
According to our reverse transcription-PCR (RT-PCR) data, Jurkat cells constitutively expressed sPLA2-IB, sPLA2-III, and sPLA2-X isozymes, whereas no transcripts of sPLA2-IIA and sPLA2-V were detectable (Figure 1). A moderate stimulation of sPLA2-IIA and sPLA2-V transcription occurred after exposure of cells to IFN-γ. The sPLA2-R was constitutively expressed and stimulated by all cytokines that were examined (Figure 1). Of note, the level of sPLA2-X mRNA decreased considerably after treatment of Jurkat cells with TNF-α.
Figure 1.
Agarose gel electrophoresis of RT-PCR amplificates of different secreted phospholipase A2 isozymes (sPLA2-IB, sPLA2-IIA, sPLA2-III, sPLA2-V, and sPLA2-X) and the M-type receptor of sPLA2 (sPLA2-R) in comparison to GAPDH mRNA in control and cytokine-primed Jurkat cells, with and without exposure to the DNA demethylating agent, 5-aza-dC. Cells were incubated for 72 hours with vehicle, 10 µM 5-aza-dC, or 5-aza-dC in combination with different proinflammatory cytokines at final concentrations of 25 ng/ml. Lane 1 in both 5-aza-dC-treated and untreated cells represents controls (without addition of cytokines) for cytokine-treated cells. Lanes 2 to 6 represent cells treated with IFN-γ, IL-1β, IL-1β + TNF-α (both at 12.5 ng/ml), TNF-α, and IL-6, respectively. Data are representative of at least three independent experiments giving similar results.
Preincubation of Jurkat cells with the methyltransferase inhibitor, 5-aza-dC, induced the transcription of sPLA2-IIA, increased existing expression of sPLA2-R, but did not significantly affect expression of sPLA2-IB, sPLA2-III, sPLA2-V, or sPLA2-X (Figure 1). The failed strong effect of 5-aza-dC treatment on sPLA2-IB, sPLA2-III, and sPLA2-X expression was reproducible using RT-PCR with lower amounts of amplification cycles (30 instead of 40 cycles to exclude a possible end point amplification; data not shown). The additional presence of proinflammatory cytokines, IFN-γ, TNF-α, IL-1β, or IL-6, was required to induce sPLA2-V by 5-aza-dC treatment (Figure 1).
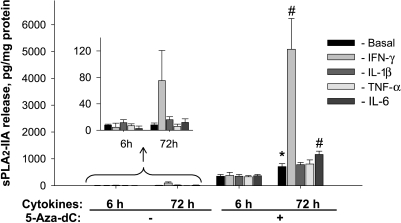
Similar results to the above at the protein level for sPLA2-IIA were obtained as determined using a commercially available ELISA kit. After exposure of Jurkat cells to IFN-γ for 72 hours, the level of sPLA2-IIA secreted into the culture medium increased significantly from 8.1 ± 2.9 to 75.1 ± 45.6 pg/mg cell protein (Figure 2). A strong up-regulation of sPLA2-IIA resulted after simultaneous treatment of cells with cytokines and 5-aza-dC (Figure 2). The sPLA2-IIA protein level released into culture medium increased from 708.4 ± 112.0 pg/mg cell protein in cells treated with 5-aza-dC for 72 hours to 5084.4 ± 1149.0 pg/mg cell protein when cells were exposed to IFN-γ and 5-aza-dC during the same period.
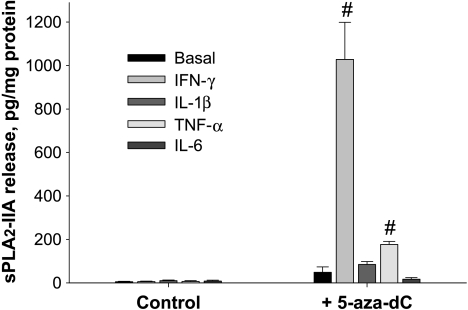
Figure 2.
Bar graphs showing effects of the demethylating agent, 5-aza-dC, alone and in combination with proinflammatory cytokines on sPLA2-IIA protein levels in conditioned culture medium of Jurkat cells. Cytokines were added at a final concentration of 25 ng/ml. Cells were exposed to 10 µM 5-aza-dC or vehicle (DMSO) for 6 and 72 hours. The insert shows at higher resolution the results for sPLA2-IIA protein release in cells that were not exposed to 5-aza-dC. Results are means ± SDs of three separate analysis in quadruplicates (n = 12). *P < .05 versus cells not exposed to 5-aza-dC; #P < .05 versus cells exposed to 5-aza-dC but not to cytokines.
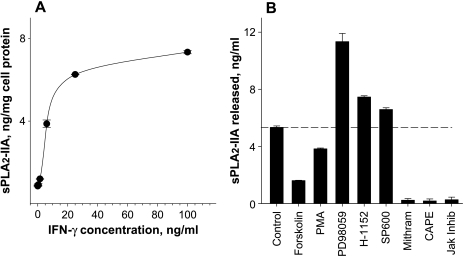
A comparison of sPLA2-IIA mRNA expression and protein release in cells treated with 5-aza-dC combined with IFN-γ indicates stronger effects on sPLA2-IIA protein release (Figure 2) than on mRNA level (Figure 1). A possible reason for this difference is that after 72 hours of incubation when the protein release and sPLA2-IIA mRNA levels were determined, the peak level of mRNA was earlier and transient, whereas the amount of sPLA2-IIA protein in the culture medium reflects the accumulation of the released enzyme during the incubation period of 72 hours. This conclusion is consistent with findings of Vial et al. [30], who observed a peak level of sPLA2-IIA mRNA at 16 hours and its decline at 32 hours, whereas the sPLA2-IIA protein level remained high until the end of incubation. Analysis of the active concentration range for IFN-γ indicated that IFN-γ-mediated up-regulation of sPLA2-IIA expression in Jurkat cells was dose-dependent, with a half-maximal effect at ∼6 ng/ml of IFN-γ (Figure 3A).
Figure 3.
Dose-dependent effects of IFN-γ (A) and effects of pharmacological manipulations of cell signaling pathways (B) on sPLA2-IIA protein release from Jurkat cells treated with 5-aza-dC. Concentrations of sPLA2-IIA protein in cell incubates determined after a 72-hour exposure of cells to 10 µM 5-aza-dC and increasing concentrations of IFN-γ are shown in panel A. Effects of cell signaling activators and inhibitors on the levels of sPLA2-IIA after a 72-hour incubation with 25 ng/ml IFN-γ and 10 µM 5-aza-dC are shown in panel B. Forskolin (10 µM), PMA (30 ng/ml), H-1152 (10 µM), PD98059 (PD98; 50 µM), indirubin (100 nM), SP-600125 (SP600; 5 µM), mithramycin A (Mithram; 250 nM), CAPE (25 µM), or Jak inhibitor I (Jak Inhib; 1 µM) were added simultaneously with IFN-γ and 5-aza-dC. The data shown are the means ± SDs of analysis in triplicate and are representative of three independent experiments.
Regulation of sPLA2-IIA Expression by the Activity of Signaling Pathways
A marked increase of sPLA2-IIA protein release occurred when 5-aza-dC- and IFN-γ-treated cells were incubated with the mitogen-activated/extracellular response protein kinase/extracellular signal-regulated kinase 1/2 inhibitor, PD-98059 (Figure 3B). Expression was also increased, albeit to a lesser extent, after incubation of 5-aza-dC- and IFN-γ-treated cells with H-1152, a Rho-kinase inhibitor, and SP-600125, a Jun N-terminal kinase inhibitor.
An inhibition of sPLA2-IIA protein release was found when Jurkat cells treated with 5-aza-dC and IFN-γ were exposed simultaneously to forskolin (Figure 3B). A similar inhibitory effect was seen by PMA as an activator of protein kinase C. Finally, mithramycin A, an inhibitor of specificity protein 1 (Sp1) binding to DNA, CAPE, an inhibitor of NF-κB, and Janus kinase inhibitor I completely abolished the reexpression of sPLA2-IIA induced by 5-aza-dC and IFN-γ in Jurkat cells (Figure 3B).
Effects of 5-aza-dC Treatment on the Methylation Status of Selected 5′-CpG Sites in the Proximal sPLA2-IIA Promoter
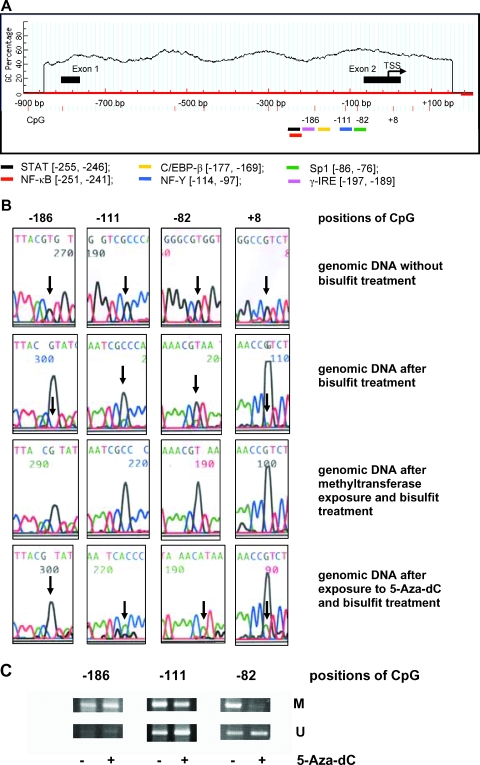
The analyses using MethPrimer demonstrated that distinct 5′-CpG islands are present only in PLA2G10 and PLA2-R genes. No 5′-CpG islands were detected in the promoter regions of PLA2G2A (Figure 4A), PLA2G1B, PLA2G3, and PLA2G5 genes (data not shown). Furthermore, using TFSEARCH and ooTFD software, we found that the sPLA2-IIA promoter region from -260 to +20 contained a number of potential binding sites, including those for signal transducer and activator of transcription (STAT) [-255, -246], NF-κB [-251, -241], CAAT-enhancer.binding protein β (C/EB-β) [-177, -169], NF-Y [-114, -97], Sp1 [-86, -76], and γ-IRE [-197, -189] (Figure 4A).
Figure 4.
Methylation of the proximal promoter of the PLA2G2A gene. (A) Location of 5′-CpG sites and potential binding regions for transcription factors in the proximal promoter of PLA2G2A gene was assessed using MethPrimer and TFSEARCH software as described in the Materials and Methods section. The gene sequence is numbered [-900, +200 bp] relative to the transcription start site [29] and the proximal promoter region [-260, +20] of PLA2G2A. Positions of potential binding sites for transcription factors, STAT, NF-κB, C/EBP-β, NF-Y, Sp1, and γ-IRE, are illustrated. (B) Methylation status of selected 5′-CpG sites in the proximal PLA2G2A promoter of Jurkat genomic DNA before and after exposure to 10 µM 5-aza-dC as described in the Materials and Methods section. Representative results of sequence analyses of the bisulfite-treated genomic reverse DNA strands are shown. Arrows indicate the conversion of reverse 5′-CpG sites, -186, -111, -82, and +8, in dependence on the methylated status. (C) Methylation-specific PCR analyses of genomic DNA before and after exposure of Jurkat cells to 5-aza-dC. M or U indicate the presence of methylated or unmethylated 5′-CpG sites.
In Figure 4B, data of sequence analyses of 5′-CpG sites -186, -111, -82, and +8 in the reverse strand are shown. A complete conversion of guanine into adenine demonstrated that the bisulfite modification was effective. Similar results were obtained by analysis of the forward strand (data not shown). As a positive control, enzymatically hypermethylated genomic DNA from Jurkat cells was used. Here, the residual unmethylated portions of 5′-CpG sites -111, -82, and +8 disappeared completely as a sign of total methylation of these sites (Figure 4B).
Finally, sequence analyses of genomic DNA from Jurkat cells demonstrated that the 5′-CpG sites -186, -111, -82, and +8 were methylated to ∼100%, ∼83%, ∼79%, and nearly 100%, respectively. After 5-aza-dC treatment for 72 hours, the methylated portions of 5′-CpG sites -111, -82, and +8 decreased to ∼20%, 0%, and ∼83%, respectively. The methylation status of the 5′-CpG site -186 did not changed significantly after exposure of cells to 5-aza-dC (Figure 4B).
The data obtained by sequence analyses were supported by methylation-specific PCR, showing that the 5′-CpG -82 site was strongly demethylated and the -111 site partially demethylated by 5-aza-dC; in contrast, no demethylation of the 5′-CpG -186 site was evident (Figure 4C).
Reexpression of sPLA2-IIA in Human U937 Leukemia Cells
As shown in Figure 5, treatment of U937 leukemia cells with 5-aza-dC restored sPLA2-IIA expression, and this was further markedly facilitated by simultaneous addition of IFN-γ. The levels of sPLA2-IIA released into the medium of these cells were distinctly lower compared with those released into the medium by Jurkat cells. The pattern of reexpression was, however, overall qualitatively similar in both leukemic cell lines, except that IL-6 increased sPLA2-IIA release in Jurkat cells, but not in U937 cells, and TNF-α increased sPLA2-IIA release in U937 cells, but not in Jurkat cells (Figures 2 and 5).
Figure 5.
Effects of 5-aza-dC alone and in combination with proinflammatory cytokines on sPLA2-IIA protein levels released by U937 leukemia cells. Cytokines were added at a final concentration of 25 ng/ml, and cells were exposed to 10 µM 5-aza-dC or vehicle (DMSO) for 72 hours, respectively. Results are the means ± SDs of three separate analysis in quadruplicates (n = 12). #P < .05 versus cells exposed to 5-aza-dC but not to cytokines.
Discussion
In this study of five secreted phospholipases A2 (groups IB, IIA, III, V, and X) in T-lymphoblastic Jurkat cells, we show that epigenetic mechanisms regulate the expression of two isozymes, sPLA2-IIA and sPLA2-V. Scanning of 5′-flanking promoter regions (-1000; +200) of corresponding PLA2G2A and PLA2G5 genes indicated a relatively low number of 5′-CpG sites and no 5′-CpG islands. In contrast, PLA2G10 and PLA2-R genes are rich in 5′-CpG islands (data not shown). However, the constitutive expression of PLA2G10 and PLA2-R and failed strong effects of 5-aza-dC on the expression of both genes argue against a significant involvement of epigenetic mechanism such as hypermethylation in the regulation of these genes in Jurkat cells.
Nevertheless, that the PLA2G2A and PLA2G5 genes have no 5′-CpG islands and the presence of clusters of methylated 5′-CpG sites or even a single site—instead of 5′-CpG islands within the binding regions of crucial transcription factors—can be sufficient for gene silencing as shown for other genes [31–33]. Such sites may also be responsible for silencing of PLA2G2A and PLA2G5.
By using TFSEARCH [23,24] and ooTFD [25] software, we confirmed a number of specific cognate binding sites in the proximal promoter region from -260 to +20 of the PLA2G2A gene for transcription factors known to be important for the PLA2G2A promoter activity, including Sp1, NF-κB, STAT, and C/EBP-β [4,18,19,29]. Two additional binding sites were found for NF-Y, also called CCAAT box-binding factor [34], and γ-interferon response element (γ-IRE) in the same region (Figure 4A). The former transcription factor, NF-Y, is considered to be important for the PLA2G2A promoter activity [18]. How far the latter transcription factor, γ-IRE, in addition to previously examined transcription factors, is actually essential for promoter activity, however, requires further investigation.
The data from sequence analyses and methylation-specific PCR demonstrated that four 5′-CpG sites (-186, -111, -82, and +8) at this sPLA2-IIA promoter region were almost completely methylated. After 5-aza-dC treatment, two of the sites (-82 and -111) became quantitatively demethylated, whereas some demethylation occurred at the +8 site and negligible demethylation at the -186 site.
The demethylation of 5′-CpG sites at -82 and -111 of the sPLA2-IIA proximal promoter regions by 5-aza-dC may be of special importance. As shown in Figure 4A, site -82 is within the potential Sp1 binding domain, 5′-GACCACGCC-3′, near the TATA box of the sPLA2-IIA promoter [18,35], whereas site -111 is part of the DNA binding region for NF-Y/CCAAT box-binding factor. Recently, it was demonstrated that binding of both transcription factors is crucial for full induction of the promoter activity of the human PHGDH gene in HeLa cells [36]. Possibly, therefore, hypermethylation of 5′-CpG sites at -82 and -111 of the proximal PLA2G2A promoter may result in failure of both transcription factors to bind at putative Sp1 and NF-Y binding sites, with subsequent absence of sPLA2-IIA expression. The importance of Sp1 binding in the regulation of sPLA2-IIA expression is supported by our findings that blockade of Sp1 binding sites by mithramycin A completely abolished the induction of sPLA2-IIA by 5-aza-dC and IFN-γ. Mithramycin A acts as an inhibitor of Sp1. It binds to 5′-CpG sites and 5′-CpG-rich tracts as a dimer that forms in the presence of magnesium [37]. The importance of NF-Y for sPLA2-IIA promoter activity, however, still has to be analyzed more thoroughly.
Our findings that the Jak inhibitor I and CAPE, an inhibitor of NF-κB, completely blocked the 5-aza-dC- and IFN-γ-mediated induction of sPLA2-IIA (Figure 3) suggest that for an optimal sPLA2-IIA expression, in addition to Sp1, the activities of NF-κB and STAT are necessary. The activities of these transcription factors are modulated by upstream cell signaling pathways. As demonstrated in a variety of cell types—from human aortic smooth muscle cells, HepG2 hepatoma cells, and prostate epithelial cells to human malignant prostate cell lines (PC-3 and LNCaP)—all of these pathways have negative regulatory influences on sPLA2-IIA expression [4,19]. Our data indicate the presence of similar negative regulatory influences on the expression of sPLA2-IIA in Jurkat cells.
Our data, however, also indicated different effects of cAMP/protein kinase A (PKA) signaling pathway on the sPLA2-IIA expression in Jurkat cells compared with other cell types [38,39]. In normal prostate epithelial cells and tumor prostate PC-3 cells, for example, activation of cAMP/PKA by forskolin in the presence of IFN-γ results in a synergistic up-regulation of sPLA2-IIA [19]. In contrast, an inhibitory effect of forskolin has been observed in malignant prostate LNCaP cells [19] and lipopolysaccharide (LPS)-stimulated alveolar macrophages [40], a response similar to the effect observed in the present study in Jurkat cells. The reason for the divergent effects of the cAMP/PKA signaling pathway among different cell types presently remains unclear. Interestingly, trichostatin A, an inhibitor of histone deacetylase activity, decreased the LPS-stimulated sPLA2-IIA expression in alveolar macrophages [40]. It is known that histone deacetylase activity is inhibited by PKA-dependent phosphorylation [41]. This suggests that histone deacetylation as a further epigenetic mechanism may be involved in the forskolin-mediated effect on the LPS-stimulated sPLA2-IIA expression in alveolar macrophages. Whether a similar mechanism is also acting in Jurkat cells remains unknown.
The importance of diverse cell signaling cascades in the regulation of sPLA2-IIA expression underscores the opportunity that, in addition to the direct demethylation of 5′-CpG sites in the PLA2G2A promoter itself, indirect mechanisms may also be responsible for the reexpression of sPLA2-IIA mediated by 5-aza-dC. It is conceivable that this may involve demethylation of 5′-CpG sites in the promoters of corresponding upstream signaling components or transcription factors. Indeed, our search of 5′-CpG islands confirmed the presence of distinct islands in gene promoters of relevant regulators of sPLA2-IIA expression, including Sp1, NF-κB p65, Jak/STAT, Rho-kinase, and C/EBP-β (data not shown).
Interferon response elements have also been identified to exhibit 5′-CpG islands [42]. Of note, the sPLA2-IIA promoter at positions [-197, -189] has a potential binding site for the γ-IRE (Figure 4A). It is interesting that, in leukemic cells, such regulatory elements were down-regulated due to hypermethylation of 5′-CpG motifs in their promoter regions [42]. Therefore, it is possible that IREs are also involved in the restoration of sPLA2-IIA expression by 5-aza-dC.
In general, the regulation of the PLA2G2A promoter is complex and includes species- and cell type-specific mechanisms of induction by proinflammatory mediators (for review [3,4,29]). TNF-α and IL-1β effectively up-regulate the expression of sPLA2-IIA in numerous cell types; moreover, a strong up-regulation of sPLA2-IIA was found solely by IFN-γ [4] including cell lines with reexpressed PLA2G2A after 5-aza-dC treatment such as malignant prostate DU-145 cells [19] or Jurkat and U-937 cells as shown here.
The observation that the inhibition of DNA methylation can restore the cytokine-mediated effects on sPLA2-IIA and sPLA2-V expression may have important implications in the development of novel strategies to treat leukemia. Cytokines are known to be crucial in a variety of cellular and molecular events [43,44]. For example, IFN-γ stimulates antiproliferative pathways in tumor cell lines [45]. Expression data in immortalized fibroblasts revealed that among genes with methylation-dependent silencing, approximately 50% were regulated through interferon-dependent growth-suppressive signaling pathways [46]. Furthermore, the efficiency of cytokinemediated effects can be amplified in cells treated with 5-aza-dC. For example, 5-aza-dC and IFN-γ at relatively low individual concentrations acted in a proapoptotic and synergistic manner in resistant neuroblastoma, medulloblastoma [47], and metastatic uveal melanoma cells [48].
Finally, our current data demonstrate that epigenetic silencing of PLA2G2A and PLA2G5 is not restricted to Jurkat cells but also occurs in leukemic U937 cells. The pathophysiological relevance of PLA2G2A and PLA2G5 silencing in malignant cells remains unclear. Nevertheless, the finding of the present study support the growing body of evidence that the antioncogenic and proapoptotic properties of sPLA2 isozymes may have importance for novel strategies targeting defective apoptosis pathways in tumors [49–53]. It should be noted, however, that 5-aza-dC treatment induces not only some well-established antioncogenic genes but also a set of prometastatic genes [54]. Therefore, establishing in detail which genes are upregulated by 5-aza-dC treatment is important to understanding the overall prooncogenic or antioncogenic effects of such a treatment.
In conclusion, the data of this study indicate that DNA methylation plays a crucial role in the regulation of sPLA2-IIA and sPLA2-V in Jurkat and U937 cells. Whether a direct hypermethylation of single 5′-CpG sites of the proximal promoter may explain sPLA2-IIA silencing in Jurkat and other cells or whether the hypermethylation of 5′-CpG sites in the promoter of corresponding upstream acting transcription factors or signaling components are responsible requires further investigation.
Acknowledgments
The authors are grateful to Margot Vogel and Sabine Fodor for technical assistance. The authors also thank Eberhard Kuhlisch for helpful discussions.
Abbreviations
- 5-aza-dC
5-aza-2′-deoxycytidine
- C/EBP-β
CAAT-enhancer-binding protein β
- CAPE
caffeic acid phenethyl ester
- DMSO
dimethyl sulfoxide
- ELISA
enzyme-linked immunosorbent assay
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IFN-γ
interferon γ
- γ-IRE
γ-interferon response element
- LPS
lipopolysaccharide
- IL-1β
interleukin 1β
- IL-6
interleukin 6
- Jak
Janus kinase
- NF-κB
nuclear factor κB
- NF-Y
nuclear factor Y
- PKA
protein kinase A
- PMA
phorbol-12-myristate 13-acetate
- RT-PCR
reverse transcription-polymerase chain reaction
- Sp1
specificity protein 1
- sPLA2-IB
secreted phospholipase A2 of group IB
- sPLA2-IIA
secreted phospholipase A2 of group IIA
- sPLA2-V
secreted phospholipase A2 of group V
- sPLA2-X
secreted phospholipase A2 of group X
- STAT1
signal transducer and activator of transcription 1
- TNF-α
tumor necrosis factor α
References
- 1.Six DA, Dennis EA. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim Biophys Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 2.Kudo I, Murakami M. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 2002;68–69:3–58. doi: 10.1016/s0090-6980(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 3.Valentin E, Lambeau G. Increasing molecular diversity of secreted phospholipase A2 and their receptors and binding proteins. Biochim Biophys Acta. 2000;1488:59–70. doi: 10.1016/s1388-1981(00)00110-4. [DOI] [PubMed] [Google Scholar]
- 4.Menschikowski M, Hagelgans A, Siegert G. Secretory phospholipase A2 of group IIA: is it an offensive or a defensive player during atherosclerosis and other inflammatory diseases? Prostaglandins Other Lipid Mediat. 2006;79:1–33. doi: 10.1016/j.prostaglandins.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Lambeau G, Gelb MH. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu Rev Biochem. 2008;77:495–520. doi: 10.1146/annurev.biochem.76.062405.154007. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita S, Ogawa M, Sakamoto K, Abe T, Arakawa H, Yamashita J. Elevation of serum group II phospholipase A2 levels in patients with advanced cancer. Clin Chim Acta. 1994;228:91–99. doi: 10.1016/0009-8981(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 7.Jiang JZ, Neubauer BL, Graff JR, Chedid M, Thomas JE, Roehm NW, Zhang S, Eckert GJ, Koch MO, Eble JN, et al. Expression of group IIA secretory phospholipase A2 is elevated in prostatic intraepithelial neoplasia and adenocarcinoma. Am J Pathol. 2002;160:667–671. doi: 10.1016/S0002-9440(10)64886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laye JP, Gill JH. Phospholipase A2 expression in tumours: a target for therapeutic intervention? Drug Discov Today. 2003;8:710–716. doi: 10.1016/s1359-6446(03)02754-5. [DOI] [PubMed] [Google Scholar]
- 9.Sved P, Scott KF, McLeod D, King NJ, Singh J, Tsatralis T, Nikolov B, Boulas J, Nallan L, Gelb MH, et al. Oncogenic action of secreted phospholipase A2 in prostate cancer. Cancer Res. 2004;64:6934–6940. doi: 10.1158/0008-5472.CAN-03-3018. [DOI] [PubMed] [Google Scholar]
- 10.Gorovetz M, Baekelandt M, Berner A, Trope' CG, Davidson B, Reich R. The clinical role of phospholipase A2 isoforms in advanced-stage ovarian carcinoma. Gynecol Oncol. 2006;103:831–840. doi: 10.1016/j.ygyno.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 11.Cummings BS. Phospholipase A2 as targets for anti-cancer drugs. Biochem Pharmacol. 2007;74:949–959. doi: 10.1016/j.bcp.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Tribler L, Jensen LT, Jørgensen K, Brünner N, Gelb MH, Nielsen HJ, Jensen SS. Increased expression and activity of group IIA and X secretory phospholipase A2 in peritumoral versus central colon carcinoma tissue. Anticancer Res. 2007;27:3179–3185. [PubMed] [Google Scholar]
- 13.Kashiwagi M, Friess H, Uhl W, Berberat P, Abou-Shady M, Martignoni M, Anghelacopoulos SE, Zimmermann A, Büchler MW. Group II and IV phospholipase A2 are produced in human pancreatic cancer cells and influence prognosis. Gut. 1999;45:605–612. doi: 10.1136/gut.45.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung SY, Chen X, Chu KM, Yuen ST, Mathy J, Ji J, Chan AS, Li R, Law S, Troyanskaya OG, et al. Phospholipase A2 group IIA expression in gastric adenocarcinoma is associated with prolonged survival and less frequent metastasis. Proc Natl Acad Sci USA. 2002;99:16203–16208. doi: 10.1073/pnas.212646299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seilhamer JJ, Pruzanski W, Vadas P, Plant S, Miller JA, Kloss J, Johnson LK. Cloning and recombinant expression of phospholipase A2 present in rheumatoid arthritic synovial fluid. J Biol Chem. 1989;264:5335–5338. [PubMed] [Google Scholar]
- 16.Roshak AK, Capper EA, Stevenson C, Eichman C, Marshall LA. Human calcium-independent phospholipase A2 mediates lymphocyte proliferation. J Biol Chem. 2000;275:35692–35698. doi: 10.1074/jbc.M002273200. [DOI] [PubMed] [Google Scholar]
- 17.Tessier C, Hichami A, Khan NA. Implication of three isoforms of PLA2 in human T-cell proliferation. FEBS Lett. 2002;520:111–116. doi: 10.1016/s0014-5793(02)02779-5. [DOI] [PubMed] [Google Scholar]
- 18.Paradon M, Salvat C, Fan Q, Bereziat G, Olivier JL. An Sp1-like 5′-GACCACGCC-3′ sequence is critical for activity of the inflammatory phospholipase A2 promoter and binds several non-zinc finger proteins. Eur J Biochem. 1998;258:113–122. doi: 10.1046/j.1432-1327.1998.2580113.x. [DOI] [PubMed] [Google Scholar]
- 19.Menschikowski M, Hagelgans A, Gussakovsky E, Kostka H, Paley EL, Siegert G. Differential expression of secretory phospholipases A2 in normal and malignant prostate cell lines: regulation by cytokines, cell signaling pathways, and epigenetic mechanisms. Neoplasia. 2008;10:279–286. doi: 10.1593/neo.07965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;3(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 21.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 22.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 23.Yutaka A. TFSEARCH: Searching Transcription Factor Binding Sites. http://www. rwcp.or.jp/papia.
- 24.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:364–370. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh D. Object-oriented transcription factors database (ooTFD) Nucleic Acids Res. 2000;28:308–310. doi: 10.1093/nar/28.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frommer ML, McDonald E, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 29.Andreani M, Olivier JL, Berenbaum F, Raymondjean M, Bereziat G. Transcriptional regulation of inflammatory secreted phospholipases A2. Biochim Biophys Acta. 2000;1488:149–158. doi: 10.1016/s1388-1981(00)00117-7. [DOI] [PubMed] [Google Scholar]
- 30.Vial D, Señorale-Pose M, Havet N, Molio L, Vargaftig BB, Touqui L. Expression of the type-II phospholipase A2 in alveolar macrophages. Down-regulation by an inflammatory signal. J Biol Chem. 1995;270:17327–17332. doi: 10.1074/jbc.270.29.17327. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Hattar J, Jiricny J. Methylation of single CpG dinucleotides within a promoter element of the Herpes simplex virus tk gene reduces its transcription in vivo. Gene. 1988;65:219–227. doi: 10.1016/0378-1119(88)90458-1. [DOI] [PubMed] [Google Scholar]
- 32.Douet V, Heller MB, Le Saux O. DNA methylation and Sp1 binding determine the tissue-specific transcriptional activity of the mouse Abcc6 promoter. Biochem Biophys Res Commun. 2007;354:66–71. doi: 10.1016/j.bbrc.2006.12.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou B, Chim CS, Zeng H, Leung SY, Yang Y, Tu SP, Lin MC, Wang J, He H, Jiang SH, et al. Correlation between the single-site CpG methylation and expression silencing of the XAF1 gene in human gastric and colon cancers. Gastroenterology. 2006;131:1835–1843. doi: 10.1053/j.gastro.2006.09.050. [DOI] [PubMed] [Google Scholar]
- 34.Matuoka K, Chen KY. Transcriptional regulation of cellular ageing by the CCAAT box-binding factor CBF/NF-Y. Ageing Res Rev. 2002;1:639–651. doi: 10.1016/s1568-1637(02)00026-0. [DOI] [PubMed] [Google Scholar]
- 35.Massaad C, Paradon M, Jacques C, Salvat C, Bereziat G, Berenbaum F, Olivier JL. Induction of secreted type IIA phospholipase A2 gene transcription by interleukin-1β. Role of C/EBP factors. J Biol Chem. 2000;275:22686–22694. doi: 10.1074/jbc.M001250200. [DOI] [PubMed] [Google Scholar]
- 36.Jun DY, Park HS, Lee JY, Baek JY, Park HK, Fukui K, Kim YH. Positive regulation of promoter activity of human 3-phosphoglycerate dehydrogenase (PHGDH) gene is mediated by transcription factors Sp1 and NF-Y. Gene. 2008;414:106–114. doi: 10.1016/j.gene.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Hampshire AJ, Fox KR. The effects of local DNA sequence on the interaction of ligands with their preferred binding sites. Biochimie. 2008 Jan 11; doi: 10.1016/j.biochi.2008.01.001. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 38.Menschikowski M, Hagelgans A, Hempel U, Siegert G. Glycogen synthase kinase-3β negatively regulates group IIA phospholipase A2 expression in human aortic smooth muscle and HepG2 hepatoma cells. FEBS Lett. 2004;577:81. doi: 10.1016/j.febslet.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 39.Menschikowski M, Hagelgans A, Heyne B, Hempel U, Neumeister V, Goez P, Jaross W, Siegert G. Statins potentiate the IFN-γ induced upregulation of group IIA phospholipase A2 in human aortic smooth muscle cells and HepG2 hepatoma cells. Biochim Biophys Acta. 2005;1733:157–171. doi: 10.1016/j.bbalip.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Raymond B, Leduc D, Ravaux L, Goffic RL, Candela T, Raymondjean M, Goossens PL, Touqui L. Edema toxin impairs anthracidal phospholipase A2 expression by alveolar macrophages. PLoS Pathog. 2007;3:1907–1917. doi: 10.1371/journal.ppat.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortmann CA, Burchert A, Holzle K, Nitsche A, Wittig B, Neubauer A, Schmidt M. Down-regulation of interferon regulatory factor 4 gene expression in leukemic cells due to hypermethylation of CpG motifs in the promoter region. Nucleic Acids Res. 2005;33:6895–6905. doi: 10.1093/nar/gki1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97:439–447. doi: 10.1111/j.1349-7006.2006.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 45.Brandacher G, Winkler C, Schroecksnadel K, Margreiter R, Fuchs D. Antitumoral activity of interferon-γ involved in impaired immune function in cancer patients. Curr Drug Metab. 2006;7:599–612. doi: 10.2174/138920006778017768. [DOI] [PubMed] [Google Scholar]
- 46.Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, Tainsky MA. Epigenetic silencing of multiple interferon pathway genes after cellular immortalization. Oncogene. 2003;22:4118–4127. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- 47.Fulda S, Debatin KM. 5-Aza-2′-deoxycytidine and IFN-γ cooperate to sensitize for TRAIL-induced apoptosis by upregulating caspase-8. Oncogene. 2006;25:5125–5133. doi: 10.1038/sj.onc.1209518. [DOI] [PubMed] [Google Scholar]
- 48.Gollob JA, Sciambi CJ. Decitabine up-regulates S100A2 expression and synergizes with IFN-γ to kill uveal melanoma cells. Clinical Cancer Res. 2007;13:5219–5225. doi: 10.1158/1078-0432.CCR-07-0816. [DOI] [PubMed] [Google Scholar]
- 49.Costa-Junior HM, Hamaty FC, da Silva Farias R, Einicker-Lamas M, da Silva MH, Persechini PM. Apoptosis-inducing factor of a cytotoxic T cell line: involvement of a secretory phospholipase A2. Cell Tissue Res. 2006;324:255–266. doi: 10.1007/s00441-005-0095-y. [DOI] [PubMed] [Google Scholar]
- 50.Lee C, Park DW, Lee J, Lee TI, Kim YJ, Lee YS, Baek SH. Secretory phospholipase A2 induces apoptosis through TNF-α and cytochrome c-mediated caspase cascade in murine macrophage RAW 264.7 cells. Eur J Pharmacol. 2006;536:47–53. doi: 10.1016/j.ejphar.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 51.Mora R, Maldonado A, Valverde B, Gutierrez JM. Calcium plays a key role in the effects induced by a snake venom Lys49 phospholipase A2 homologue on a lymphoblastoid cell line. Toxicon. 2006;47:75–86. doi: 10.1016/j.toxicon.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Petrovic U, Sribar J, Matis M, Anderluh G, Peter-Katalinić J, Krizaj I, Gubensek F. Ammodytoxin, a secretory phospholipase A2, inhibits G2 cell-cycle arrest in the yeast Saccharomyces cerevisiae. Biochem J. 2005;391:383–388. doi: 10.1042/BJ20050417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Putz T, Ramoner R, Gander H, Rahm A, Bartsch G, Bernardo K, Ramsay S, Thurnher M. Bee venom secretory phospholipase A2 and phosphatidylinositol-homologues cooperatively disrupt membrane integrity, abrogate signal transduction and inhibit proliferation of renal cancer cells. Cancer Immunol Immunother. 2007;56:627–640. doi: 10.1007/s00262-006-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ateeq B, Unterberger A, Szyf M, Rabbani SA. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia. 2008;10:266–278. doi: 10.1593/neo.07947. [DOI] [PMC free article] [PubMed] [Google Scholar]