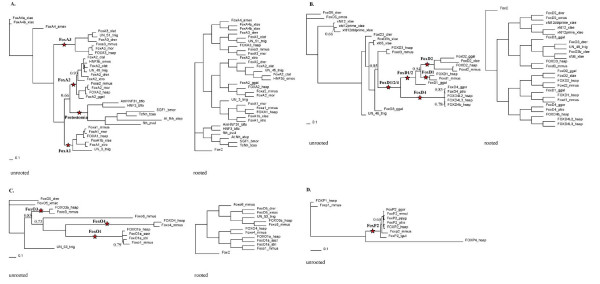
Figure 1.
Branches tested for positive selection using branch-site models. In each phylogeny the branches tested are indicated with red stars and labels representing the clade of interest. The trees in which positive selection was identified were rooted using a set of FoxC genes as an outgroup in conjunction with neighbor joining tree creation, after branch-site analysis for clarity into evolutionary relationships. Clade credibility values less than 0.95 are indicated on the unrooted trees. The phylogenies are for the gene clusters as follows; A. FoxA B. FoxD C. FoxO D. FoxP.