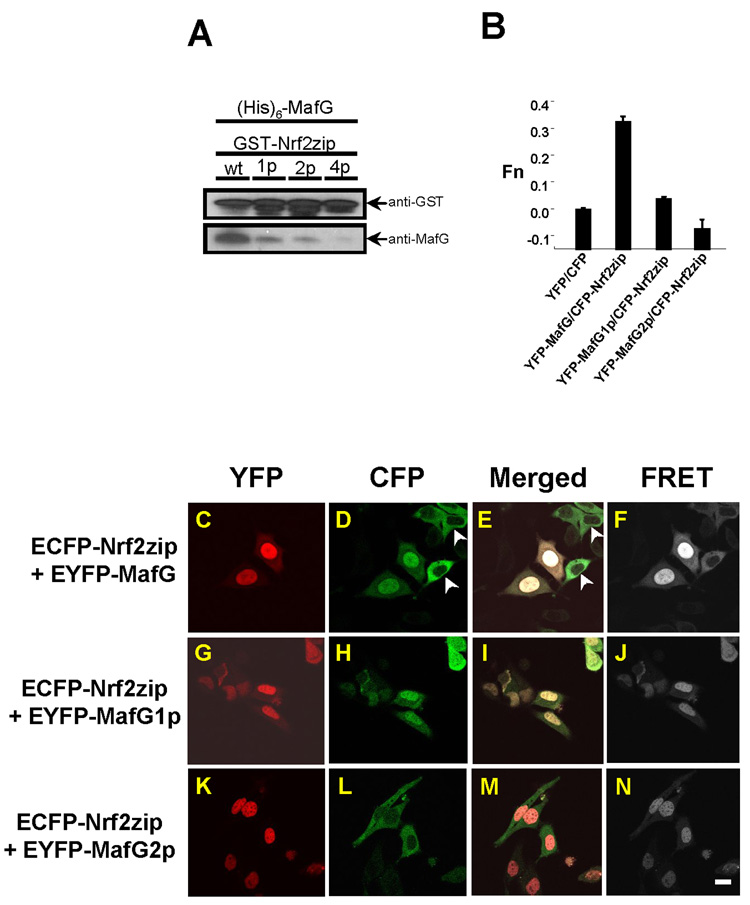
Fig. 3.
Dimerization with MafG enhances nuclear retention of Nrf2. (A) GST pull down study showed that wild type Nrf2zip exhibited the strongest binding to MafG. Single point (1p) mutation in Nrf2zip attenuated MafG binding. Two point (2p) mutation further decreased MafG binding. Four point (4p) mutation completely abolished MafG binding. (B) Calculated FRET value showed strong interaction between Nrf2zip/MafG. FRET value was attenuated in Nrf2zip/MafG1p and completely negated in Nrf2zip/MafG2p. As a negative control, CFP/YFP failed to elicit FRET signal. (C–N) Confocal microscopy and FRET assay of MafG/Nrf2zip binding. ECFP-Nrf2zip showed co-localization with EYFP-MafG and EYFP-MafG1p in the nucleus. In the absence of EYFP-MafG, ECFP-Nrf2zip exhibited a cytosolic distribution (arrowheads) (D–E). ECFP-Nrf2zip however, showed a discrete cytosolic distribution, un-overlapped with the nuclear location of EYFP-MafG2p. To enhance visual effect, the EYFP, ECFP and FRET signals are artificially represented with red, green and white color, respectively. Scale bar: 10 µm.