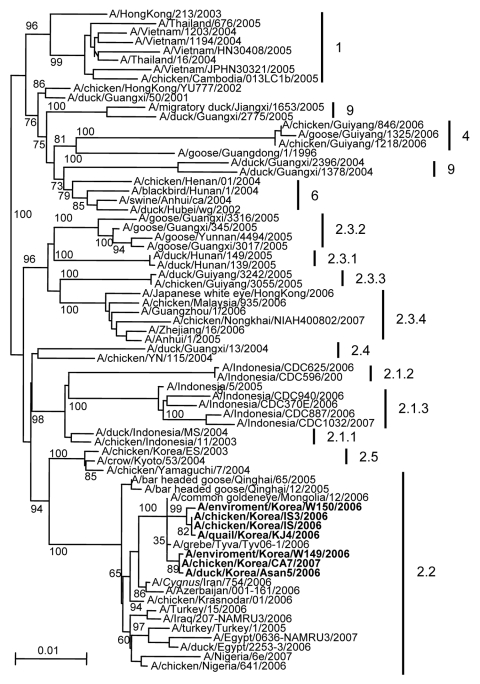
Figure 2.
Phylogenetic trees for hemagglutinin (HA) genes of Korean influenza virus (H5N1) isolates from wild birds and poultry farms during 2006–2007. The DNA sequences were compiled and edited by using the Lasergene sequence analysis software package (DNASTAR, Madison, WI, USA). Multiple sequence alignments were made by using ClustalX (10). Rooted phylograms were prepared with the neighbor-joining algorithm and then plotted by using NJplot (11). Branch lengths are proportional to sequence divergence and can be measured relative to the scale bar shown (0.01-nt changes per site). Branch labels record the stability of the branches >1,000 bootstrap replicates. The tree was produced by referring to the proposed global nomenclature system for influenza virus (H5N1) (www.offlu.net). Boldface indicates isolates tested in the current study.