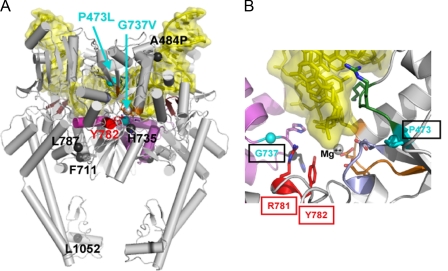
FIGURE 12.
Localization of amino acids changed in mAMSA-hypersensitive mutants. Structures shown are representations of the structure solved by Dong and Berger of the breakage-reunion domain of yeast Top2 bound to a nicked double-stranded DNA. A shows the entire breakage reunion domain from a side view. Only one of two subunits has the relevant amino acids marked. The two mutants hypersensitive to mAMSA alone (P473L and G737V) are marked in cyan; other changed residues are marked in black. The active site tyrosine (Tyr782) is indicated in red. B is a closer view of the catalytic core, including the active site tyrosine, Arg781 (critical for catalysis), Pro473, and Gly737. Other relevant structural features include the amino acid sequence PLRGK (amino acids 473–477 of yeast Top2, shown in green) EGDSA (amino acids 449–453 of yeast Top2, shown in periwinkle), and the a4 helix (shown in lavender). This helix carries several amino acids changed in other mutants conferring hypersensitivity, including Ser740 and Thr744). DNA in both illustrations is shown in yellow. It should be emphasized that the DNA shown in the illustrations are protein DNA co-crystals as solved by Dong and Berger. Mg++ indicates an electron density that is likely a Mg2+ ion.