Abstract
Little is known about the mechanisms by which the lung epithelial progenitors are initially patterned and how proximal-distal boundaries are established and maintained when the lung primordium forms and starts to branch. Here we identified a number of Notch pathway components in respiratory progenitors of the early lung, and we investigated the role of Notch in lung pattern formation. By preventing γ-secretase cleavage of Notch receptors, we have disrupted global Notch signaling in the foregut and in the lung during the initial stages of murine lung morphogenesis. We demonstrate that Notch signaling is not necessary for lung bud initiation; however, Notch is required to maintain a balance of proximal-distal cell fates at these early stages. Disruption of Notch signaling dramatically expands the population of distal progenitors, altering morphogenetic boundaries and preventing formation of proximal structures. Our data suggest a novel mechanism in which Notch and fibroblast growth factor signaling interact to control the proximal-distal pattern of forming airways in the mammalian lung.
The mammalian respiratory system arises from the ventral foregut endoderm. Studies in mice show that at around embryonic day 9 (E9)2 respiratory progenitors are collectively identified in the ventral foregut as a group of endodermal cells expressing Titf1 (thyroid transcription factor-1, or Nkx2.1). Besides being the earliest known marker of respiratory progenitors, Titf1 appears to be required to regulate lung epithelial cell fate (1). By E9.5, major morphogenetic changes occur in the respiratory field, leading to Fgf10 (fibroblast growth factor 10)-mediated expansion of these progenitors to form the lung primordia. Once primary lung buds have formed, lateral buds arise at stereotyped positions from these tubules (the main bronchi) and undergo branching morphogenesis and differentiation to give rise to the bronchial tree (2, 3).
Little is known about the mechanisms by which the lung epithelial progenitors are expanded and patterned into proximal and distal compartments during the early stages of lung morphogenesis. Furthermore, it is still unclear how proximal-distal boundaries are established and maintained when lung epithelial buds form and start to branch. Studies in other foregut derivatives, such as the pancreas and stomach implicate signaling by Notch as a critical regulator of pattern during organ formation (4, 5).
The Notch pathway orchestrates a highly evolutionarily conserved mechanism of control of cell fate decisions, which plays a prominent role in establishing asymmetries or differences in signaling between two cells. During development, Notch-mediated mechanisms give rise to cellular diversity while also serving to generate compartments and to establish tissue boundaries (6-8). Notch signaling results from cell-cell contact via interactions of Notch receptors (in mammalians, Notch1 to -4) with ligands (Delta-like 1, 3, and 4 and Jagged1 and -2) in adjacent cells. Ligand-receptor engagement results in shedding of the Notch extracellular domain and subsequent activation of a second proteolytic cleavage by a γ-secretase complex. The Notch intracellular domain (NICD) is then released and reaches the nucleus, where it associates with the transcription factor CSL (in mice, Rbpjk) and its transcriptional complex, leading to activation of the Rbpjk-mediated transcription of Hes and Hey genes. Alternatively, Notch signaling may be activated by a ligand-independent noncanonical pathway, which is still poorly understood in mammals (7-9).
Notch has been implicated in several aspects of lung biology, including a role in epithelial growth and differentiation and in carcinogenesis (10, 11). In the adult lung and in lung epithelial cell lines derived from tumors, the Notch pathway appears to function as an oncogenic or tumor suppressor signal, depending on the cellular context (12). During development, Notch receptors and ligands have been identified in both epithelial and mesenchymal compartments of the lung, and there is in vitro evidence that Notch may act on epithelial differentiation and branching morphogenesis (13, 14). Mice that are genetically deficient in Hes1, an effector of Notch signaling in several biological systems, show airway pulmonary neuroendocrine cell hyperplasia and a decreased number of secretory cells (15). Mice expressing a constitutively active Notch3 transgene in the distal lung epithelium using a surfactant protein promoter-enhancer show immature lungs in which epithelial differentiation into type I and II cells is impaired (16). It is currently unknown, however, whether Notch signaling influences cell fate decisions or morphogenetic processes in respiratory progenitors at the onset of lung development and when airway start to form. Overall the mouse genetic models available have not shed light into this problem, since deletion of key components of the Notch pathway, such as Notch1, Notch2, Jagged1, Dll1, Dll4, or Rbpjk, results in early embryonic lethality prior to or when the lung starts to develop (17-22). Although several Notch conditional mouse models are available, it is still unclear whether the existing Cre deleter lines are able to effectively inactivate these genes in lung endodermal precursors early enough to study these issues (23, 24). Moreover, the analysis of individual Notch mutants can be complicated by the possibility of functional redundancy, given that multiple Notch receptors, ligands, or effector molecules are often co-expressed.
To circumvent some of the problems above, here we used a classical pharmacological approach to disrupt global Notch signaling in the foregut and in the early lung and study the resulting phenotype. We provide evidence that components of the Notch pathway are already present in respiratory progenitors of the primary bud and that disruption of Notch signaling dramatically expands the population of distal lung progenitors, altering morphogenetic boundaries and proximal-distal lung patterning. These changes are accompanied by altered distribution of Notch components and reveal a previously unsuspected role for Notch-Jagged-Delta signaling in the lung. Our data support a mechanism in which Notch and Fgf10 interact to control cell fate and patterning of forming airways in the mammalian lung.
EXPERIMENTAL PROCEDURES
Foregut Cultures—Experiments were performed in a foregut culture system as previously described (25). Briefly, foreguts were isolated from E8.5 CD1 mouse embryos (Charles River Laboratories) in phosphate-buffered saline (PBS) using tungsten needles. Explants were cultured for 3-4 days on Costar 6-well Transwell-Col plates containing 1.5 ml of BGJb medium (Invitrogen), 10% fetal bovine serum (Invitrogen), penicillin/streptomycin, and 20 mg/dl ascorbic acid (Sigma). Cultures were incubated at 37 °C in 95% air and 5% carbon dioxide. Medium was changed daily. Once harvested, explants were either frozen for biochemical analysis or fixed in 4% paraformaldehyde for whole mount in situ hybridization (WMISH) or immunohistochemistry.
Lung Organ Cultures—E11.5-E12 lungs from CD1 mouse embryos were cultured in air-liquid interface up to 48 h on BGJb medium (Invitrogen; 25 mg/dl ascorbic acid, 1% inactivated fetal bovine serum). In some experiments, heparin beads soaked in either buffer (PBS, control) or human recombinant FGF10 (100 μg/ml; R&D Systems) were engrafted near distal buds, and lungs were cultured for 24-48 h, as described previously (26). Human recombinant FGF10 has been shown by multiple studies to effectively induce an Fgf-mediated chemoattractant response in mouse lung epithelium (25, 26, 31).
Disruption of γ-Secretase Activity in Vitro—Foregut and lung explants were treated with N-(S)-phenyl-glycine-t-butyl ester (DAPT) (Sigma) in DMSO diluted in medium to final concentrations ranging from 1 to 50 μm, depending on the experiment. Control explants consisted of medium containing DMSO (1 μl/ml). In addition, we tested three other γ-secretase inhibitors: JLK6 (5, 10, and 25 μm) (27), L685458 (L6; 1, 10, 25, and 50 μm) (28), and Compound 1 (C1; 25, 50, and 100 μm) (4).
Inactivation of E-cadherin Function in Lung Cultures—E11.5 lung explants were cultured for 48 h with the anti-E-cadherin monoclonal antibody DECMA-1 (100 μg/ml; U3254; Sigma), a well characterized blocking antibody (29, 30). Control medium contained sodium azide (50 μm).
Morphometric Analysis—The number of distal buds (including the peripheral and lateral buds) in the developing airways of lung explants was counted in control and DAPT-treated lung cultures at 4, 20, 24, 28, and 48 h (n = 6 in each group). The data (means ± S.E.) were represented in a graphic form (distal buds × culture time). Statistical analysis was performed by Student's t test (SPSS 12.0 software; SPSS, Chicago, IL), and differences (control versus DAPT) were considered significant at p < 0.05. For the quantitative analysis of cell proliferation, the number of Ki67-labeled epithelial and mesenchymal cells was counted in five nonoverlapping fields near distal buds in control and DAPT-treated lung cultures at 48 h. Cells were defined by the presence of a nucleus in tissue sections. The data (mean ± S.E.) were expressed as a percentage of total cells counted per field and analyzed by Student's t test using SPSS 12.0 software (SPSS, Chicago, IL).
Whole Mount in Situ Hybridization—Digoxigenin-labeled riboprobes (Maxiscript kit; Ambion) were generated from plasmids carrying cDNAs for Titf1, Bmp4, Sox2, and Sfptc, as previously reported (25). The cDNA templates were generated by PCR with primers containing either T7 or T3 promoter sequences (Table 1). Generation of riboprobes (sense and antisense) and whole mount in situ hybridization of freshly isolated embryonic lungs, cultured foreguts, or cultured lungs were performed as previously described (31). The specificity of all probes was demonstrated in preliminary experiments or in previous reports. No sense signal was detected in tissues in which a defined pattern of expression was obtained with the antisense probe. All conclusions were based on results from more than 3 specimens/condition.
TABLE 1.
PCR primers (forward, top; reverse, bottom) used to generate cDNA templates for in situ hybridization and PCR product size
| Primers | size | |
|---|---|---|
| bp | ||
| Notch1 | 5′-AATTAACCCTCACTAAAGGGTTACCACTCCAGCCACAGAA-3′ | 540 |
| 5′-TAATACGACTCACTATAGGGAGCATCCTGAAGCACTGGAA-3′ | ||
| Notch2 | 5′-AATTAACCCTCACTAAAGGGGTGTATCTGTTACACAGCAGT-3′ | 481 |
| 5′-TAATACGACTCACTATAGGGTTCCTGGACACTACACAGGT-3′ | ||
| Notch3 | 5′-AATTAACCCTCACTAAAGGGGTCTGCTCAATCCTGTAGCT-3′ | 520 |
| 5′-TAATACGACTCACTATAGGGAGTCATAGAGGAGTGTCACT-3′ | ||
| Notch4 | 5′-AATTAACCCTCACTAAAGGGTCACTGGGACCTGCTAACGCT-3′ | 518 |
| 5′-TAATACGACTCACTATAGGGCAGACTCGTACGTGTCGCTT-3′ | ||
| Dll1 | 5′-AATTAACCCTCACTAAAGGGAACCTCGTTCGAGACCTCAA-3′ | 538 |
| 5′-TAATACGACTCACTATAGGGGTGCGTGCTTCCTATATGAA-3′ | ||
| Dll3 | 5′-AATTAACCCTCACTAAAGGGGCACTCAACAACCTGAGGTT-3′ | 461 |
| 5′-TAATACGACTCACTATAGGGCAGGTGGATCTCTGTGAGTT-3′ | ||
| Dll4 | 5′-AATTAACCCTCACTAAAGGGAGGCTTGCAAAGGATAGGCCT-3′ | 411 |
| 5′-TAATACGACTCACTATAGGGATCGGCTTCAAGGTTGGTCT-3′ | ||
| Jag1 | 5′-AATTAACCCTCACTAAAGGCTTTCCTGCTTTAGACTTGAA-3′ | 468 |
| 5′-TAATACGACTCACTATAGGCACTAATAGAACAAACACCAA-3′ | ||
| Jag2 | 5′-AATTAACCCTCACTAAAGGGTGCTGTGCAGCGTGTTCAGT-3′ | 570 |
| 5′-TAATACGACTCACTATAGGGAGAAACTATGCACACGGCCT-3′ | ||
| RPBjk | 5′-AATTAACCCTCACTAAAGGTCGCAGACTGTACCACTGTAA-3′ | 388 |
| 5′-TAATACGACTCACTATAGGTGTTCTCAGTTATCGACTTAA-3′ | ||
| Lfng | 5′-AATTAACCCTCACTAAAGGGGAAGCTGATTTGGGTAGTAA-3′ | 631 |
| 5′-TAATACGACTCACTATAGGGCCTATGCAACAACACATCAA-3′ | ||
| Psen1 | 5′-AATTAACCCTCACTAAAGGGCCCTTACTGTCCAGGAGGTT-3′ | 568 |
| 5′-TAATACGACTCACTATAGGGCTTTGGGCACACATCCTATT-3′ | ||
| Hes1 | 5′-AATTAACCCTCACTAAAGGGTTCAGCGAGTGCATGAACGA-3′ | 568 |
| 5′-TAATACGACTCACTATAGGGGTCCGTCAGAAGAGAGAGGT-3′ | ||
| Hes5 | 5′-AATTAACCCTCACTAAAGGGTCGTTCCTCTGGATGTGGGAA-3′ | 488 |
| 5′-TAATACGACTCACTATAGGGAGCGCTCATCAGACAGCCAA-3′ | ||
| Hey1 | 5′-AATTAACCCTCACTAAAGGGATGGACCGAGGTGTTGTATA-3′ | 410 |
| 5′-TAATACGACTCACTATAGGGGTTTCACAGGCACCAAGCTA-3′ | ||
| Hey2 | 5′-AATTAACCCTCACTAAAGGGGGCTCATTGACACCAACTCAA-3′ | 469 |
| 5′-TAATACGACTCACTATAGGGTTTCTCCACACAGCAGAGAAA-3′ |
Western Blot Analysis—Western blot analysis was carried out in cultured foreguts or lungs using the following antibodies: anti-cleaved Notch1 (NICD; rabbit polyclonal antibody; Cell Signaling), anti-E-cadherin (mouse monoclonal antibody clone 36; BD Transduction, San Jose, CA), anti-β-catenin (mouse monoclonal antibody; BD Transduction), anti-panactin (mouse monoclonal; NeoMarkers, Fremont, CA), and α-tubulin (mouse monoclonal antibody; Sigma), as described (32-34). ECL Plus reagents (Amersham Biosciences) and appropriate secondary antibodies (Bio-Rad) were used for Western blotting detection.
Quantitative Real Time RT-PCR—Total RNA was isolated from cultured lungs, treated with DNase (Ambion, Austin, TX), and reverse-transcribed using Superscript II (Invitrogen). Quantitative real time PCR was performed using an ABI 7000 instrument (Applied Biosystems, Foster City, CA) and primers (Hes1, Hes5, Hey1, Hey2, Fgf10, and β-actin) obtained from Assays-on-Demand (Applied Biosystems, Austin, TX). Reactions were performed in 50 μl using TaqMan PCR universal Master (Applied Biosystems). The relative concentration of RNA for each gene to β-actin mRNA was determined using the expression, 2-ΔCT, where ΔCT = (CTmRNA - CTβ-actin RNA).
Immunohistochemistry—Immunohistochemistry was performed in 5-μm paraffin sections using an anti-Titf1 antibody (mouse monoclonal antibody; Dako Corp.) (35) and the Cell and Tissue Staining Kits (CTS Series; R&D Systems, Minneapolis, MN), according to the manufacturer's protocol.
For whole mount immunohistochemistry, lung explants (n = 3-5) were fixed with 4% paraformaldehyde for 2 h, washed with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature and blocked with 1% bovine serum albumin with 10% goat serum at 4 °C overnight. Primary antibodies (anti-E-cadherin and Ki-67; all mouse monoclonal antibodies from BD Transduction (36, 37)) were added in 1% bovine serum albumin/PBST for 6 h at room temperature. Explants were incubated with secondary antibody (goat anti-mouse antibody labeled with fluorescein; Molecular Probes) in PBST for 1 h and subsequently with rhodamine phalloidin and nuclear stain (Molecular Probes) for 30 min. Negative controls consisted of sections incubated with normal mouse IgG. Samples were mounted in VectaShield and analyzed in a Zeiss confocal laser-scanning microscope (LSM510 META).
Mesenchymal Lung Cell Cultures and Real Time PCR—Mouse neonatal lung mesenchymal (MLg) cells were cultured in control medium (Dulbecco's modified Eagle's medium, 10% fetal calf serum) or in DAPT-containing (10 or 50 μm) media for 48 h. Total RNA was isolated from cell cultures (Trizol; Invitrogen), reverse transcribed (1 μg of RNA), and amplified by real time PCR (see above), as described by Chen et al. (42).
RESULTS
Notch Pathway Components Are Present in Respiratory Progenitors of the Early Lung—We analyzed the spatial-temporal distribution of Notch components using WMISH in foreguts and lungs isolated from E9-E12.5 CD1 mouse embryos. Although Notch signaling components have been previously reported in the developing lung (15, 38-40), little is known about their expression pattern during the initial stages of lung formation.
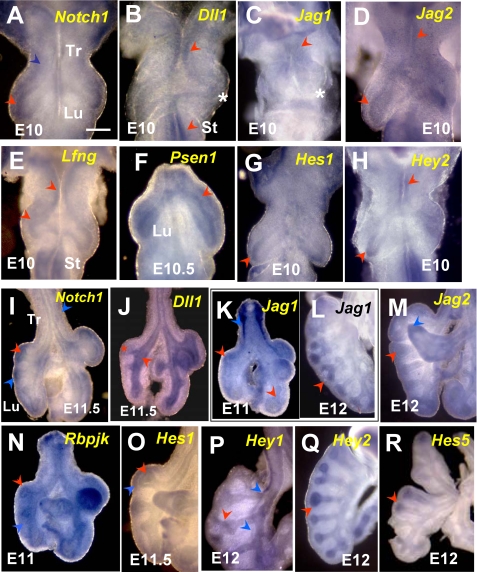
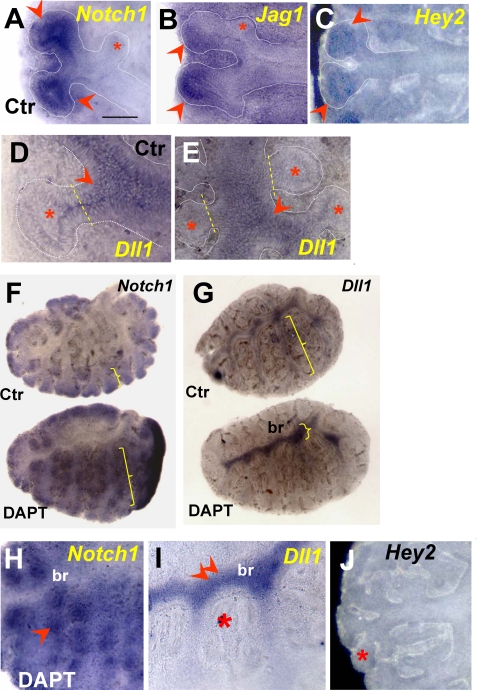
At around E9, when lung progenitors are already present in the foregut endoderm but budding has not yet initiated, endodermal expression of Notch receptors and ligands in the prospective lung field was undetectable. By E10, Notch1 transcripts were clearly present in the lung epithelial progenitors at the tips of primary buds and in the mesenchyme associated with the lung and tracheal primordia (Fig. 1A). The Notch1-expressing regions of the E10 lung epithelium also expressed Jag2 but no other Jag or Dll1 (Delta-like 1) ligands; epithelial signals for Notch2 and Notch3 were undetectable, but low levels of Notch4 appeared to be present (Fig. 1, B-D) (data not shown). The presence of Notch1 but not Notch2 and -3 in the E10 mouse lung was in agreement with RT-PCR data from a previous report (13). The expression of critical pathway components, including Lfng (Lunatic Fringe), Psen1 (Presenilin 1), and putative Notch transcriptional targets Hes1, Hey2, and Hes5 in the distal regions of the primary lung buds strongly suggested local activation of Notch signaling (Fig. 1, E-H) (data not shown). Although not present in the early lung epithelium, Jag1 was transcribed in the E10 mesenchyme, in the developing vasculature (Fig. 1C). Dll1 signals were found in the endoderm of the tracheoesophageal region and the stomach primordium, which also expressed Jag2, Lfng, and Hey2, but not Notch receptors. Dll3 expression in the lung was ubiquitously low at E10 and later (data not shown), whereas Dll4 expression has been shown by others to be restricted to the lung vasculature (41).
FIGURE 1.
WMISH of Notch pathway components in mouse E10-10.5 foreguts (A-H) and E11-12 lungs (I-R). A-H, at E10-E10.5, strong Notch1, Hes1, and Hey2 transcripts are present in the distal epithelium of primary lung buds. These transcripts overlap with Jag2, Lfng, and Psen1 mRNAs, which are also present in the epithelium of the proximal lung, tracheoesophageal region (Tr), and prospective stomach (St). At E10, Dll1 signals cannot be identified in the lung but are present in the epithelium of the prospective trachea and stomach. Jag1 is expressed in the mesenchyme associated with the trachea and proximal lung but not in the epithelium. Notch1, Psen1, and Lfng are also transcribed in the mesenchyme of the E10-E10.5 lung and trachea. I-R, during initiation of branching, Notch1 mRNA is expressed in the epithelium of E11-E12 distal buds, which then also express Jag1, Jag2, Hey1, Hey2, Hes1, and Hes5 transcripts. From around E11 onward, Dll1 is expressed in the proximal lung epithelium and is excluded from the bud tips. The E11-E12 lung mesenchyme expresses Notch1, Jag1, Jag2, Hey1, and Hes1. Rbpjk is ubiquitously expressed at all times. The red and blue arrowheads depict epithelial and mesenchymal expression, respectively; asterisks represent undetectable signals; n = 3-5 specimens/condition. Bar (A), 250 μm.
At E11-E11.5, Notch1 and Jag2 mRNAs continued to be detected in the distal epithelial tubules (Fig. 1, I and M), which then started to express Jag1 (Fig. 1K) (39, 40). Proximal airways expressed Dll1 but showed little to no Notch or Jag mRNA (Fig. 1, I-L). The Notch transcriptional effector Rbpjk was expressed ubiquitously (Fig. 1N), and the Notch target genes Hes1, Hes5, and Hey2 were detected in the airway epithelium predominantly in the distal buds (Fig. 1, O, Q, and R). This suggested that epithelial activation of Notch occurred predominantly at the tips of branching tubules. Hey1 signals were present at low levels in the epithelium but were strong in lung vascular structures (Fig. 1P). In agreement with previous reports, we also found Jag1, Jag2, Notch1 to -4, Hey1, Hes1, and Dll4 mRNAs strongly expressed in the E11-E12 mesenchyme, where they could potentially play a role in the development of the lung vasculature or other derivatives, such as smooth muscle or cartilage (Fig. 1) (data not shown).
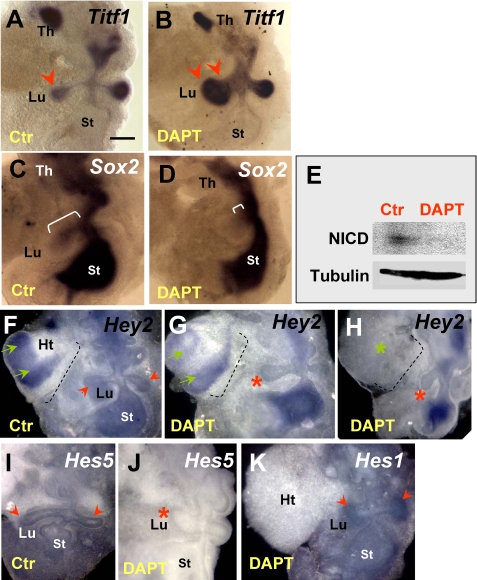
Notch Signaling Restricts Expansion of Distal Progenitors during Primary Lung Bud Formation—We asked whether Notch activity was required in respiratory progenitors for proper formation of the lung primordium. To circumvent problems related to potential functional redundancy and early embryonic lethality reported in Notch genetic models, we used a pharmacological approach to disrupt global Notch signaling in vitro in the foregut at the onset of lung development. We have previously reported that foregut explants isolated from E8.5 mouse embryos can give rise to different derivatives in vitro and can be used as a model system to study lung bud initiation (25, 42). Thus, we investigated the role of Notch in primary lung bud formation by analyzing foregut explants growing under control conditions (DMSO-containing medium) or in the presence of DAPT (Sigma). This compound has been widely reported to inhibit γ-secretase-mediated cleavage of Notch, blocking the function of all Notch receptors in a variety of developing systems (4, 43-48). Preliminary studies in foregut or lung organ cultures (see below) suggested that we could inhibit Notch signaling at DAPT concentrations ranging from 1 to 50 μm (Figs. 2E and 3, A and B) (data not shown). In the control foreguts, the lung buds are typically identified by Titf1, a marker of early thyroid and lung progenitors, which continues to be expressed in the lung epithelium in a proximal-distal gradient, with higher levels distally (Fig. 2A). Treatment with DAPT (25-50 μm) as early as E8.5 did not prevent induction of primary lung buds. The lung primordium, however, appeared truncated in its proximal region while markedly enlarged distally. The strong Titf1 expression in the large buds suggested an abnormal accumulation of distal epithelial progenitors, resulting in altered proximal-distal patterning of the lung primordium (Fig. 2B). This was further supported by analysis of Sox2, an SRY-related HMG box transcription factor, expressed in the proximal lung epithelium. As reported by others and shown in our controls, Sox2 is transcribed in the endoderm of the developing foregut derivatives, including the lung. During bud formation, Sox2 labels proximal, but not distal, morphogenetically active epithelium (Fig. 2C) (49, 50). Fig. 2, C and D, shows a dramatic reduction in the domain of Sox2 mRNA expression in DAPT-treated foreguts, consistent with the truncation of the proximal region of the lung primordium. The phenotype was highly penetrant and was observed in the vast majority of the explants (>95%, n > 20). Other foregut derivatives looked grossly unaltered (data not shown). We had evidence that Notch signaling was active in controls and was indeed blocked in the foreguts presenting this abnormal phenotype. Efficacy of blocking was systematically tested by Western blot analysis of these explants using an antibody known to recognize the γ-secretase-cleaved Notch1 fragment (Val1744 epitope) but not the uncleaved receptor (33). As shown in Fig. 2E, an NICD band was consistently detected in control but not in DAPT-treated foregut cultures (Fig. 2E). Disruption of Notch signaling was further confirmed by down-regulation of the putative Notch target genes Hey2 and Hes5 mRNAs in lung buds from DAPT-treated explants (Fig. 2, F-J). By contrast, Hes1 expression was apparently not affected by DAPT treatment, although the lung buds were clearly abnormal. This strongly suggested that Hey2 and Hes5, but not Hes1, are potential candidate mediators of the Notch effects in the primary lung bud. Interestingly, although local Hey2 expression was inhibited by DAPT in the lung epithelium (see also the heart field), signals were preserved elsewhere in the foregut endoderm (Fig. 2, F-H). Furthermore, the severity of the lung phenotype appeared to correlate with the overall degree of Hey2 down-regulation in DAPT-responsive tissues. For example, in Fig. 2, F-H, the lung bud is most truncated in the explant in which Hey2 transcripts were nearly undetectable in the lung field (also compare Hey2 expression in the heart field in Fig. 2, F-H).
FIGURE 2.
Inhibiting γ-secretase activity in the foregut disrupts proximal-distal pattern of the lung primordium. A and B, WMISH of Titf1 in control foregut cultures (E8.5 + 3 days) shows strong signals in lungs buds (Lu) distally (arrowhead) and in the thyroid primordium (Th). DAPT treatment (50 μm) of foreguts results in expansion of the Titf1-expressing lung region (double arrowheads) and shortening of the proximal region. C and D, in controls, Sox2 labels the foregut endoderm, including the proximal lung (bracket), which is markedly truncated in DAPT-treated explants (small bracket). E, Western blot of cleaved Notch (NICD) shows loss of expression in DAPT-treated foreguts. F-H, WMISH of Hey2 in control foregut culture shows strong signals in heart (Ht) and in the foregut endoderm, including the primary lung buds. G and H, in DAPT-treated foreguts, Hey2 expression is selectively abolished in heart and lung buds but not elsewhere in the foregut. The severity of the lung phenotype correlates with the overall degree of Hey2 down-regulation in DAPT-responsive tissues (compare signals in the heart; green arrows). I and J, Hes5 is present in the lung and stomach regions of the control foregut, and expression is abolished by DAPT. However, Hes1 expression in distal lung buds seems unchanged after DAPT treatment (K) (data not shown). n = 3-6 foreguts/condition in both WMISH and Western blot experiments. Bar (A), 300 μm.
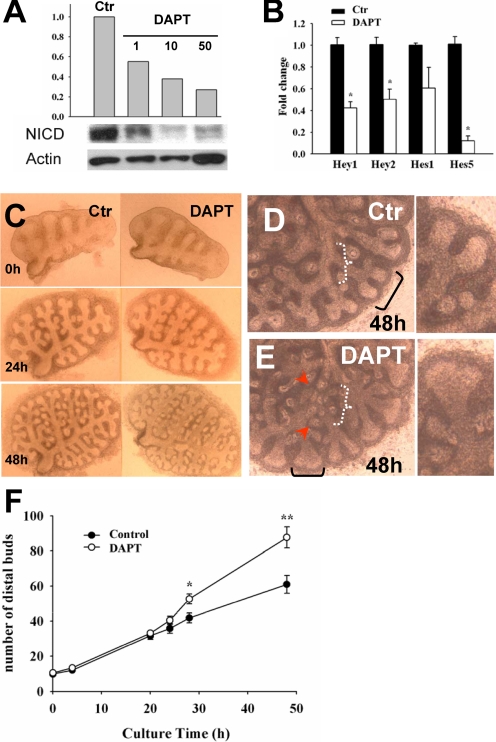
FIGURE 3.
DAPT inhibits Notch signaling and induces ectopic budding in cultured lungs. A, representative Western blot shows dose-dependent inhibition of cleaved Notch1 (NICD) in E11.5 lungs treated with DAPT (1-50 μm) for 48 h. The histogram depicts relative values (densitometry) of NICD normalized by β-actin in control (DMSO) and DAPT groups. B, real time PCR shows Hey1, Hey2, and Hes5 but not Hes1 significantly down-regulated by DAPT in cultured lungs (-fold change, mean ± S.E.; n = 3, p < 0.05). C-E, branching pattern of cultured lungs (control, DAPT). By 48 h, DAPT-treated lungs show dilated terminal buds (black brackets in D and E) and multiple ectopic buds arising in more proximal regions (E; depicted by arrowheads within white bracket). F, the histogram represents the number of distal buds (terminal and lateral buds) per time (h) in culture (mean ± S.E.; n = 6 in each group; *, p < 0.05; **, p < 0.01).
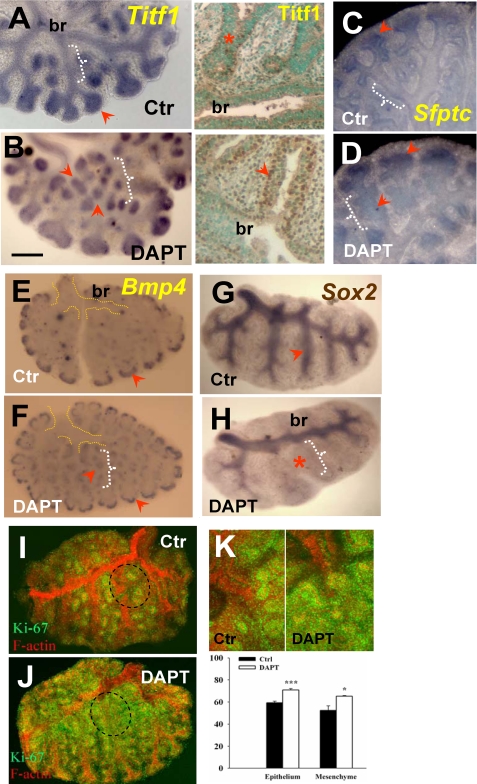
Disruption of γ-Secretase Activity Alters Morphogenetic Boundaries and Results in Ectopic Budding—The observations above suggested that in epithelium of the lung primordium Notch signaling ensures the proper balance of proximal-distal cell fates and prevents disproportionate expansion of the distal progenitors. We asked whether Notch had a similar role during initiation of branching by analyzing E11.5 lungs cultured under control (DMSO) or DAPT conditions. First, we used Western blot analysis (Val1744 epitope) to show that NICD was present in control lungs and that we could inhibit γ-secretase cleavage of Notch over a range of DAPT concentrations in lung cultures (Fig. 3A). Moreover, we found by quantitative RT-PCR analysis significant down-regulation of Notch downstream targets Hes5, Hey1, and Hey2 but not Hes1 in DAPT-treated lungs, compared with controls (Fig. 3B; see also WMISH below). Time lapse analysis of the pattern of growth of these explants revealed no obvious morphological differences between the groups before 24 h (n > 10). By 48 h, however, DAPT-treated lungs consistently showed dilated terminal buds, reminiscent of the phenotype observed in the foregut culture (Fig. 3, C-E). Remarkably, DAPT also induced a large number of buds ectopically in more proximal regions. Quantitative analysis of the number of distal buds showed an average of 87 ± 6 (mean ± S.E.) buds in DAPT-treated lungs (n = 6) versus 61 ± 5 buds in controls at 48 h (n = 6) (Fig. 3F). WMISH of DAPT-treated lungs showed that the dilated terminal buds and the ectopic buds expressed marker genes normally found in distal lung progenitors, such as Titf1, Bmp4 (bone morphogenetic-4), and Sfptc (surfactant protein c) (Fig. 4, A-F) (3). Additional proof of altered lung proximal-distal patterning was provided by the dramatic reduction in the domain of Sox2 mRNA expression, consistent with our observations in the foregut (Fig. 4, G and H). Sox2 expression seemed preserved in the epithelium of proximal regions that had already formed prior to DAPT exposure (e.g. the main bronchi; Fig. 4H). This suggested that DAPT does not act by respecifying cell fate in preexisting airways, but it was influencing proximal-distal identity of the airways that formed later in culture. Assessment of cell proliferation by Ki-67 staining (51) revealed diffuse increased labeling in both epithelial and mesenchymal compartments of DAPT-treated lungs. This is illustrated in Fig. 4, I and J, in which explants were double-stained for F-actin (to facilitate visualization of the lung structure) and Ki-67. Quantitative analysis further confirmed an increased proportion of Ki-67 labeling in distal buds of DAPT-treated lungs as compared with controls (n = 4/group; Fig. 4K). Together, these data suggested that DAPT-induced changes in proximal-distal patterning not only involved changes in cell fate but also derepression of a cell proliferation program in the distal lung.
FIGURE 4.
Disruption ofγ-secretase activity expands distal epithelial cell fate and increases distal proliferation during branching. WMISH of Titf1 (A), Sfptc (C), and Bmp4 (E) in control lungs at 48 h shows increased or selective expression of these genes in the epithelium of distal buds (arrowhead) and little to no expression in proximal epithelium (white brackets). These distal marker genes are ectopically expressed in proximal regions of DAPT-treated (50 μm) lungs (arrowhead near white bracket in B, D, and F). Immunostaining of Titf1 (A and B, right) confirms strong ectopic expression in the proximal region of DAPT-treated lungs. G and H, WMISH of Sox2 shows typical expression in proximal epithelium of controls and marked reduction in the Sox2 domain by DAPT (note preserved expression in main bronchi (br)). I and K, immunohistochemistry analysis of Ki-67 (green) and F-actin (red) in controls shows Ki-67 labeling associated mostly with budding regions (terminal and lateral buds; circled area). In DAPT-treated lungs, Ki-67 was increased where ectopic buds formed (compare the F-actin and Ki-67 expression in proximal regions in I and J). J, the increased diffuse Ki67 labeling in the most superficial layer of the proximal mesenchyme partially obscures F-actin signals. K, quantitative analysis of Ki-67-positive cells in budding regions of control and DAPT groups (upper panels, representative field) shows increased labeling in both epithelium and mesenchyme of DAPT (graph shows the percentage of positive cells among total cells; mean ± S.E.; n = 4; *, p < 0.05; **, p < 0.01); all plates are representative of n > 3 explants. Bar (B), 400 μm.
Integrity of Notch Signaling Is Required for Proper Establishment of Notch1-Dll1 Domains in the Lung Epithelium—We investigated how loss of γ-secretase activity in cultured lung explants altered expression of the Notch components previously identified in the epithelium of E11-E12 lungs. Analysis of controls at 24 and 48 h revealed Notch1, Jag1, Jag2, and Hes-Hey transcripts in the distal epithelium of branching tubules, consistent with the pattern previously seen in E11-E12 lungs (Figs. 1 and 5). Dichotomous branching was associated with down-regulation of these Notch-related genes in the interbud region (Fig. 5, A-C). Moreover, none of these genes could be detected during the initial stages of formation of lateral buds (e.g. see asterisks in Fig. 5, A and B). Expression of Dll1 was confined to nonbranching regions of the airway epithelium, and Dll1 down-regulation in emerging new buds seemed to precede induction of Notch1 and Jag genes in newly formed lateral buds (Fig. 5, D and E). DAPT treatment expanded the domains of Notch1 expression, to encompass the majority of the epithelial tubules (Fig. 5, F and H). Although highly expressed, Notch1 was nonfunctional, as also described in another Notch loss of function model, the Pofut1 (protein O-fucosyltransferase 1) null mice (52), and suggested here by the down-regulation of Hey2 mRNA (Fig. 5J). By contrast, the domain of Dll1 expression in DAPT-treated lungs was greatly reduced and restricted to the main bronchi (Fig. 5, G and I). Interestingly, at this region Dll1 signals seemed to be stronger and more uniformly expressed in the main bronchi of DAPT-treated lungs than in controls. Presumably, under normal conditions, very low levels of Notch signaling may still be present in proximal airways to control Dll1 locally. At a later developmental stage Dll1 expression has been reported in isolated clusters of airway epithelial cells identified as pulmonary neuroendocrine cells (12, 39).
FIGURE 5.
Notch1-Dll1 expression domains are markedly altered by DAPT in cultured lungs. In control 48 h cultured lungs (A-E and upper panels in F and G), Notch1, Jag1, and Hey2 transcripts are highly expressed in the epithelium at the tips of bifurcating buds (arrowheads) but not in the emerging lateral buds (asterisks). Dll1 expression is excluded from the tips of bifurcating buds and from the emerging lateral buds (D and E) but is expressed in the stalks and more proximal epithelium (arrowheads). In 48 h DAPT-treated lungs (F and G (bottom) and H-J), the domain of Notch1 expression is markedly expanded to proximal regions, whereas the Dll1 domain becomes restricted mostly to the main bronchi (compare brackets in F and G, asterisk in I), where Dll1 signals appear up-regulated (double arrowhead in I). Disruption of Notch signaling by DAPT is suggested by the loss of Hey2 expression at the tips (compare J and C). n = 3-5 explants/condition. Bar (A), 200 μm.
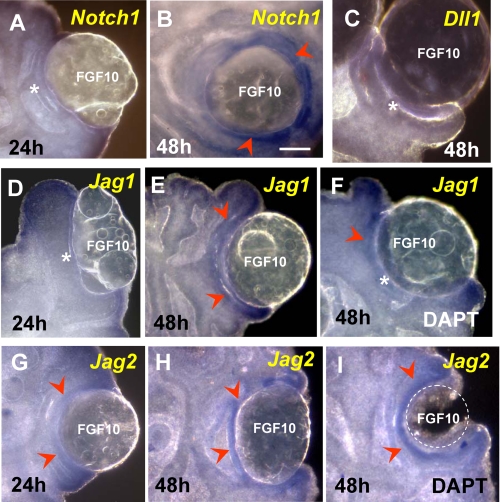
Fgf10 Induces Expression of Notch1 and Jag Ligands, but Not Dll1, in Developing Lungs—Studies in the developing pancreas, tooth, and other organs have shown that activation of Fgf signaling can induce expression of components of the Notch pathway to influence epithelial cell behavior (4, 53, 54). We asked whether this could be also true in the developing lung epithelium, since we found Notch ligands and receptors induced at the sites where Fgf10/Fgfr2b signaling is known to be most active. To address this question, we increased local Fgf activity in the lung epithelium using recombinant FGF10 and looked for changes in expression of Notch-related genes. FGF10-soaked heparin beads and control buffer (PBS)-containing beads were engrafted onto E11.5 lungs and subsequently cultured for 24-48 h (26). WMISH showed that neither Notch1 nor Jag1 was induced by FGF10 at 24 h (Fig. 6, A and D). Low levels of Jag2 could be seen in some explants at 24 h (Fig. 6G). However, by 48 h, these genes were up-regulated in the epithelium adjacent to the FGF10 bead (Fig. 6, B, E, and H). By contrast, Dll1 was not induced by FGF10 at any time point (Fig. 6C). Engraftment of control PBS beads had no effect on gene expression (not shown). Similar experiments in DAPT-treated lungs showed that FGF10-mediated induction of Jag1 but not Jag2 or Notch1 was attenuated in the epithelium (Fig. 6, F and I) (data not shown). Together, the data suggest a role for Fgf10 in the establishment of Notch receptor-ligand expression in the lung epithelium. It also suggests that once Notch signaling is established, this pathway contributes to maintain proper levels of Jag1 expression in distal buds.
FIGURE 6.
Expression of Notch1, Jag1, and Jag2, but not Dll1, is induced in the lung epithelium by exogenous FGF10. Shown is an assessment of Notch1, Dll1, Jag1, and Jag2 gene induction (WMISH) by recombinant FGF10 in beads engrafted onto E11.5 lungs and cultured for 24-48 h. At 24 h, only Jag2 showed some low level of inducibility in the epithelium associated with the FGF10-containing bead (arrowhead in G; compare with A and D). By 48 h, Notch1, Jag1, and Jag2 were highly induced by FGF10 (arrowheads in B, E, and H), whereas Dll1 was not induced at any time (C). DAPT treatment attenuates the FGF10-mediated induction of Jag1 (F) but not of Jag2 (I) or Notch1 (not shown). n = 3-4 explants/condition. Bar in B, 200 μm.
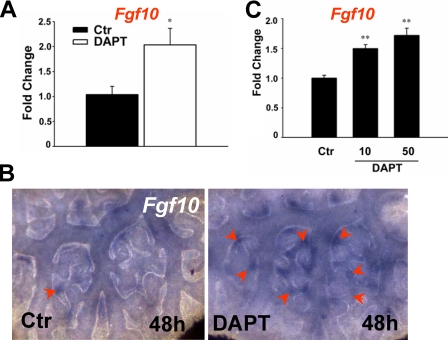
Suppression of Fgf10 Expression by Notch Signaling—To find whether the effect of Notch inactivation on ectopic bud formation was accompanied by changes in Fgf10 expression, we examined levels and distribution of this gene in control and DAPT-treated lungs by quantitative RT-PCR and WMISH. RT-PCR revealed a significant increase in Fgf10 mRNA levels in DAPT-treated lungs (Fig. 7A); WMISH confirmed the overall up-regulation of Fgf10 and showed several foci of Fgf10-expressing cells in the mesenchyme associated with the ectopic buds of DAPT-treated lungs (Fig. 7B). We asked whether Fgf10 expression was changed in a per cell basis and resulted from disruption of Notch signaling selectively in the mesenchymal cells. To address this issue, we used the lung mesenchyme-derived MLg cells, which have been previously used to study transcriptional regulation of Fgf10 (42) and express all Notch receptors (not shown). Real time PCR analysis of control and DAPT-treated MLG cells showed a dose-dependent increase in Fgf10 expression by DAPT at 48 h (Fig. 7C). Together, the data suggested that Notch signaling in the lung mesenchyme may contribute to lung patterning by exerting a repressive role in Fgf10 expression.
FIGURE 7.
DAPT up-regulates Fgf10 expression in cultured lungs and in lung mesenchyme-derived MLg cells. A, real time PCR shows up-regulation of Fgf10 mRNA in 48 h DAPT-treated lungs. B, WMISH reveals increased Fgf10 expression in the proximal mesenchyme associated with the ectopic buds of DAPT-treated lungs (arrowheads; 48 h). C, real time PCR shows dose-dependent up-regulation of Fgf10 mRNA in DAPT-treated MLg cells. Bars and lines represent mean and S.E. of at least three experiments; *, p < 0.05 (Student's t test).
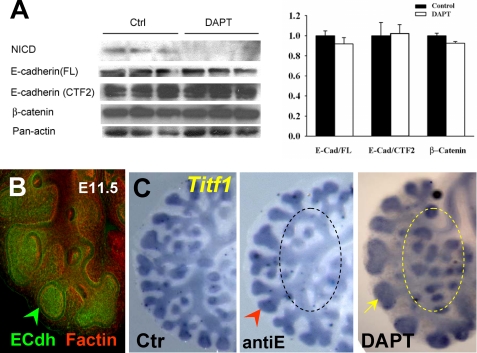
Potential Role of Additional Signals in the Phenotype from DAPT-treated Lungs—Proteolytic processing by γ-secretase has been reported in several type I membrane proteins besides Notch, including APP, ErbB4, LRP, E-cadherin, Nectin1-α, and CD44 (55). We asked whether impaired function of other γ-secretase substrates could be contributing to generate the DAPT-induced phenotype described here. A literature survey revealed that except for E-cadherin, these molecules were either not expressed in the developing lung epithelium at the stages we analyzed, or if present, they were not shown to be required for proximal-distal lung patterning in vitro or in vivo (56-61). In the developing lung epithelium, high levels of E-cadherin were found in the distal regions undergoing active branching (Fig. 8B). The effect of disrupting E-cadherin selectively in the lung is currently unknown, and E-cadherin-null mice die early in development (59). We tested whether the phenotype observed in DAPT-treated lungs was also associated with inhibition of γ-secretase-mediated cleavage of E-cadherin. For this, we used an antibody raised against the cytoplasmic region of E-cadherin (C36; BD Biosciences), which recognizes the full-length and cleaved fragments, including the γ-secretase-mediated cleaved E-cadherin (34). Western blot analysis of DAPT-treated lungs (50 μm) in which NICD expression was markedly inhibited showed no difference in the pattern of staining or intensity of the E-cadherin compared with controls (Fig. 8A). Furthermore, there were no significant changes in β-catenin expression (anti-β-catenin antibody; BD Biosciences) that suggested major alterations in E-cadherin-β-catenin complexes due to DAPT (Fig. 8A). Moreover, culturing lung explants with a well characterized anti-E-cadherin blocking antibody (anti-E (100 μg/ml); U3254; Sigma) (30) inhibited branching but did not result in the changes in proximal-distal patterning seen in DAPT-treated lungs (compare the morphology and Titf1 staining depicted by the area circled in anti-E and -DAPT panels of Fig. 8C).
FIGURE 8.
The phenotype of DAPT-treated lungs is not associated with inhibition ofγ-secretase cleavage of E-cadherin. A, Western blot analysis of NICD, E-cadherin (C36 antibody), β-catenin, and panactin in 48 h cultured lungs in which Notch cleavage was blocked by DAPT (50 μm). There is no difference in the pattern of staining or intensity of the full-length E-cadherin (E-Cad/FL), E-cadherin fragment potentially cleaved by γ-secretase (E-Cad/CTF2), or β-catenin. The graph represents densitometry measurements of Western blots (mean ± S.E.; values normalized by panactin; n = 3/group). B, whole mount immunostaining of E-cadherin (green) and F-actin (red) in E12 lung shows strongest E-cadherin signals in distal buds. C, WMISH of Titf1 in lungs cultured in control, E-cadherin blocking antibody (antiE; 100 μg/ml), and DAPT (50 μm)-containing media (48 h). AntiE inhibits branching (arrowhead) but does not result in the enlargement of peripheral buds (yellow arrow) or ectopic budding (circled area) characteristic of DAPT-treated lungs. n = 3/condition.
Finally, we compared the DAPT effects in the lung with that of JLK6, an isocoumarin-based serine protease known to inhibit γ-secretase activity of other substrates but not of Notch (27). Analysis of NICD, Sox2, and Titf1 expression confirmed that Notch cleavage was preserved and that proximal-distal patterning was undisturbed in JLK6-treated explants (5-25 μm) (supplemental Fig. 1). Treatment with two other γ-secretase inhibitors known to interfere with Notch cleavage (L685458 and Compound 1) (4, 28) did not significantly alter expression of NICD even at nearly toxic high concentrations. Interestingly, consistent with the presence of NICD, no changes in patterning or expression of Sox2 and Titf1 were observed in these lungs (supplemental Fig. 1). Together, these results further suggest a link between the phenotype we described and the disruption of γ-secretase cleavage of Notch.
DISCUSSION
Here we have identified critical components of the Notch pathway in the respiratory field of the foregut when the lung primordium is forming. We provide evidence that Notch components are expressed in respiratory progenitors and that γ-secretase cleavage of Notch is not required for primary lung bud formation. Our findings, however, reveal a novel role for Notch in balancing proximal-distal epithelial cell fate and patterning of the early lung.
Results from our expression profiling suggested that Notch1 is the predominant Notch receptor expressed in putative distal lung progenitors at the stages analyzed here. Although our functional assay targeted all Notch receptors, we had direct evidence that Notch1 cleavage was blocked in lungs that showed an abnormal phenotype. Thus, it is likely that Notch1 may play a role in mediating the Notch effects in the early lung epithelium. We cannot rule out, however, the contribution of other Notch receptors present in the lung epithelium presumably at lower levels compared with Notch1. Along these lines, Notch3 has been reported in the lung predominantly in the mesenchyme but also at low levels in the epithelium of branching tubules (39). Interestingly, Notch1 antisense oligonucleotide treatment has been reported to increase the number of branch points in peripheral buds of cultured mouse lungs (13). Here we also found an overall increase in branching in DAPT-treated lungs; however, the hallmark feature of this phenotype was the presence of ectopic distal buds in proximal regions. Discrepancies between the Notch antisense and DAPT phenotypes could be potentially ascribed to DAPT interfering with multiple Notch family members or to the disruption of an additional, yet uncharacterized, γ-secretase-dependent signal besides Notch.
Our results support the idea that Notch acts in the early lung epithelium via downstream targets that include Hey2 and Hes5 but not Hes1. Hes1 has been implicated by genetic data in specification of endocrine versus nonendocrine pulmonary cell fate. It is less clear, however, whether Notch is upstream of Hes1 in the developing lung (15). We provide evidence that disruption of Notch function in the lung epithelium at early developmental stages has a relatively minor effect in Hes1 expression.
Genetic studies in the Drosophila tracheal system strongly suggest that Notch signaling controls the number of cells destined to assume a distal position in the forming primary bud (62-64). Disruption of Notch signaling in Drosophila mutants results in accumulation of large clusters of epithelial cells at distal positions of the primary bud. This phenotype is strikingly reminiscent of the phenotype we have observed in the lung primordium of DAPT-treated foreguts. Furthermore, the ectopic budding we reported in DAPT-treated lungs has been also described in Drosophila mutants in which reduction in Notch function produces ectopic terminal branches in the developing trachea (64).
How does Notch influence proximal-distal patterning in forming airways? From the present study, Notch appears to exert this effect by influencing multiple processes, including epithelial cell fate and cell proliferation. We found that Notch restricts proliferation of tip cells. Supporting evidence includes our analysis of Ki-67 expression and the fact that Notch1 signaling appears in the lung epithelium at a relatively late stage of bud outgrowth. The involvement of Notch in cell cycle regulation and in modulating the proliferative responses of cells to other signals has been demonstrated in different biological systems (8, 65). For example, in the mouse developing inner ear, Notch-Jag1 signaling limits proliferation of prosensory cells by enabling expression of the cell cycle inhibitor Cdkn1b in these cells (66). During murine retinal angiogenesis, Dll4/Notch signaling prevents VEGF-mediated overproliferation of tip endothelial cells and ensures normal vascular patterning (67). Our results are consistent with the idea that Notch activation in the developing distal lung limits Fgf10-mediated proliferation of distal progenitor cells to maintain a proper ratio of tips and stalk cells during branching.
Fgf-Notch interactions appear to be, at least in part, functionally conserved during development of the murine and fly respiratory tract. In both species, Fgf (Branchless or Fgf10) induces budding and also induces expression of Notch components. In Drosophila, Fgf induces Dll expression in cells that occupy the tips of branching tubules. Dll activation of Notch signaling in neighbor proximal cells then prevents these cells from acquiring a distal (tip) cell phenotype (62, 64). A fundamental difference between the Drosophila and the mammalian systems is that, in the murine lung, we found that Fgf induces Jag instead of Dll and that Dll1 expression is excluded from the tips. Although both Jag ligands are Fgf10 targets in the lung epithelium, we found differences in how their expression is regulated. When compared with Jag1, Jag2 appears to be induced earlier (in primary buds and in response to recombinant FGF10). Also, in contrast to Jag1, we found that Jag2 is not dependent on Notch signaling for maintenance.
How did DAPT-induced changes in the lung mesenchyme contribute to the phenotype described here? Our Ki-67 and gene expression data support a role for Notch signaling in regulating levels and number of Fgf10-expressing cells in the lung mesenchyme. DAPT treatment appears to derepress this mechanism and create ectopic Fgf10 centers, which then induce ectopic buds. Interestingly, the ectopic tracheal budding seen in Drosophila Notch null mutants is also accompanied by an increased number of Fgf (bnl)-expressing cells, as well as increased intensity of bnl mRNA (62). Thus, the mechanism by which Notch influences bud formation may be at least in part through acting in Fgf10-producing mesenchymal cells. Investigating precisely how Notch exerts this function was beyond the scope of the present study.
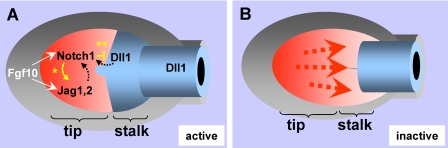
Our data suggest that during lung bud formation, epithelial cells with the highest Fgf activity lose Dll1 and later on gain Notch1 and Jag1 and -2 expression (see model in Fig. 9). We propose that distal progenitors exposed to high Fgf10 levels activate a Jag-Notch-mediated program that is dependent on Fgf signaling to initiate and is in part maintained at the tips by a mechanism compatible with lateral induction. As described in other systems, through this mechanism, local activation of Notch signaling maintains ligand production in neighbor cells, which results in a uniform field of Notch activation (66, 68, 69). This is in agreement with the increased expression of Notch1, Jag1, and Jag2 found in distal buds and with the attenuation of Jag1 induction by FGF10 in DAPT-treated lungs. Activation of Notch signaling at the tips, however, does not assign or maintain the distal phenotype in lung buds. As demonstrated in Fig. 4, tip cells from nascent lung buds already express distal markers (Titf1 and Sfptc) prior to the onset of Notch signaling. Furthermore, expression of Titf1 or Sfptc is not inhibited by DAPT.
FIGURE 9.
Proposed mechanism of Notch regulation of proximal-distal patterning during lung bud morphogenesis. A, distal epithelial progenitors (in red) exposed to local high levels of Fgf10 activate an Fgf10-Fgfr2b program that results in cell expansion and subsequent induction of Notch1 and Jag ligands. Notch signaling is maintained at the tips by Fgf and in part by Notch-mediated induction of Jag1 in neighbor cells (*, lateral induction), which contributes to establish a uniform field of Notch activation distally. Activation of Jag-Notch signaling restricts further expansion of distal progenitors. In the forming stalk, Notch1 interaction with Dll1 at the tip-stalk boundary inhibits Notch expression proximally (**, lateral inhibition) to allow stalk cells to assume a proximal phenotype. Together, these ensure a proper ratio of tip versus stalk cells and proper proximal-distal patterning of the lung. Patterning is also influenced by mesenchymal Notch signaling via a control of levels and distribution of Fgf10 (mechanism not explored here and thus not included in this diagram). B, disruption of Notch signaling results in uncontrolled expansion of distal progenitors and altered proximal-distal patterning of the lung epithelium in which stalk cells assume a distal phenotype.
Our model also predicts that in the developing lung bud, as endogenous Fgf10 becomes less available to activate signaling in cells away from the tips, fewer Jag ligands but more Dll1 is present to interact with Notch1 in the epithelium near the stalks. We propose that Dll1-Notch interaction down-regulates Notch1 expression in forming stalks, probably via lateral inhibition, to allow these cells to assume a proximal phenotype. One of the most striking features of the DAPT-treated lungs is the failure to induce typically proximal genes, such as Sox2 and Dll1 in the stalk regions of the ectopic buds, which are otherwise morphologically indistinguishable from a normal bud. This is consistent with the idea that γ-secretase activation of Notch is ultimately required for proper development of proximal epithelial structures in the lung. In the developing kidney, a similar role has been described for Notch in regulating proximal nephron cell fate (43). In summary, our study supports a role for Notch signaling in regulating patterning and proximal-distal cell fate in the early lung.
Supplementary Material
Acknowledgments
We thank Jun Qian and Fengzhi Shao for excellent technical assistance. We also thank Sheede Khalil and Mihai Nita-Lazar for help with Western blotting and the immunostaining, and Andrew McMahon, Parviz Minoo, Tom Gridley, and Olivier Pourquie for cDNA clones. We are grateful to Sue Menko (Thomas Jefferson University) for making the confocal/imaging facility available.
This work was supported, in whole or in part, by National Institutes of Health, NHLBI, Grant PO1 HL47049 (to W. V. C.) and NIH/EE Grant 014798 (to J. W. and M. A. K.). This work was also supported by National Health Research Institutes-Taiwan Physician Scientist Award PS9402 (to P. N. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: En, embryonic day n; DAPT, N-(S)-phenyl-glycine-t-butyl ester; NICD, Notch intracellular domain; PBS, phosphate-buffered saline; WMISH, whole mount in situ hybridization; RT, reverse transcription.
References
- 1.Minoo, P., Su, G., Drum, H., Bringas, P., and Kimura, S. (1999) Dev. Biol. 209 60-71 [DOI] [PubMed] [Google Scholar]
- 2.Maeda, Y., Dave, V., and Whitsett, J. A. (2007) Physiol. Rev. 87 219-244 [DOI] [PubMed] [Google Scholar]
- 3.Cardoso, W. V., and Lu, J. (2006) Development 133 1611-1624 [DOI] [PubMed] [Google Scholar]
- 4.Miralles, F., Lamotte, L., Couton, D., and Joshi, R. L. (2006) Int. J. Dev. Biol. 50 17-26 [DOI] [PubMed] [Google Scholar]
- 5.Murtaugh, L. C., Stanger, B. Z., Kwan, K. M., and Melton, D. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14920-14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis, J. (1998) Semin. Cell Dev. Biol. 9 583-589 [DOI] [PubMed] [Google Scholar]
- 7.Radtke, F., Schweisguth, F., and Pear, W. (2005) EMBO Rep. 6 1120-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artavanis-Tsakonas, S., Rand, M. D., and Lake, R. J. (1999) Science 284 770-776 [DOI] [PubMed] [Google Scholar]
- 9.Ilagan, M. X., and Kopan, R. (2007) Cell 128 1246. [DOI] [PubMed] [Google Scholar]
- 10.Dang, T. P., Gazdar, A. F., Virmani, A. K., Sepetavec, T., Hande, K. R., Minna, J. D., Roberts, J. R., and Carbone, D. P. (2000) J. Natl. Cancer Inst. 92 1355-1357 [DOI] [PubMed] [Google Scholar]
- 11.Konishi, J., Kawaguchi, K. S., Vo, H., Haruki, N., Gonzalez, A., Carbone, D. P., and Dang, T. P. (2007) Cancer Res. 67 8051-8057 [DOI] [PubMed] [Google Scholar]
- 12.Collins, B. J., Kleeberger, W., and Ball, D. W. (2004) Semin. Cancer Biol. 14 357-364 [DOI] [PubMed] [Google Scholar]
- 13.Kong, Y., Glickman, J., Subramaniam, M., Shahsafaei, A., Allamneni, K. P., Aster, J. C., Sklar, J., and Sunday, M. E. (2004) Am. J. Physiol. 286 L1075-L1083 [DOI] [PubMed] [Google Scholar]
- 14.Shan, L., Aster, J. C., Sklar, J., and Sunday, M. E. (2007) Am. J. Physiol. 292 L500-L509 [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., Udaka, N., Yazawa, T., Okudela, K., Hayashi, H., Sudo, T., Guillemot, F., Kageyama, R., and Kitamura, H. (2000) Development 127 3913-3921 [DOI] [PubMed] [Google Scholar]
- 16.Dang, T. P., Eichenberger, S., Gonzalez, A., Olson, S., and Carbone, D. P. (2003) Oncogene 22 1988-1997 [DOI] [PubMed] [Google Scholar]
- 17.Swiatek, P. J., Lindsell, C. E., del Amo, F. F., Weinmaster, G., and Gridley, T. (1994) Genes Dev. 8 707-719 [DOI] [PubMed] [Google Scholar]
- 18.Hamada, Y., Kadokawa, Y., Okabe, M., Ikawa, M., Coleman, J. R., and Tsujimoto, Y. (1999) Development 126 3415-3424 [DOI] [PubMed] [Google Scholar]
- 19.Hrabe de Angelis, M., McIntyre, J., 2nd, and Gossler, A. (1997) Nature 386 717-721 [DOI] [PubMed] [Google Scholar]
- 20.Xue, Y., Gao, X., Lindsell, C. E., Norton, C. R., Chang, B., Hicks, C., Gendron-Maguire, M., Rand, E. B., Weinmaster, G., and Gridley, T. (1999) Hum. Mol. Genet 8 723-730 [DOI] [PubMed] [Google Scholar]
- 21.Oka, C., Nakano, T., Wakeham, A., de la Pompa, J. L., Mori, C., Sakai, T., Okazaki, S., Kawaichi, M., Shiota, K., Mak, T. W., and Honjo, T. (1995) Development 121 3291-3301 [DOI] [PubMed] [Google Scholar]
- 22.Gale, N. W., Dominguez, M. G., Noguera, I., Pan, L., Hughes, V., Valenzuela, D. M., Murphy, A. J., Adams, N. C., Lin, H. C., Holash, J., Thurston, G., and Yancopoulos, G. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15949-15954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris, K. S., Zhang, Z., McManus, M. T., Harfe, B. D., and Sun, X. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2208-2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, K. S., Korfhagen, T. R., Bruno, M. D., Kitzmiller, J. A., Wan, H., Wert, S. E., Khurana Hershey, G. K., Chen, G., and Whitsett, J. A. (2007) J. Clin. Invest. 117 978-988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desai, T. J., Malpel, S., Flentke, G. R., Smith, S. M., and Cardoso, W. V. (2004) Dev. Biol. 273 402-415 [DOI] [PubMed] [Google Scholar]
- 26.Park, W. Y., Miranda, B., Lebeche, D., Hashimoto, G., and Cardoso, W. V. (1998) Dev. Biol. 201 125-134 [DOI] [PubMed] [Google Scholar]
- 27.Petit, A., Pasini, A., Alves Da Costa, C., Ayral, E., Hernandez, J. F., Dumanchin-Njock, C., Phiel, C. J., Marambaud, P., Wilk, S., Farzan, M., Fulcrand, P., Martinez, J., Andrau, D., and Checler, F. (2003) J. Neurosci. Res. 74 370-377 [DOI] [PubMed] [Google Scholar]
- 28.Wang, X. D., Leow, C. C., Zha, J., Tang, Z., Modrusan, Z., Radtke, F., Aguet, M., de Sauvage, F. J., and Gao, W. Q. (2006) Dev. Biol. 290 66-80 [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa, M., Fukata, M., Yamaga, M., Itoh, N., and Kaibuchi, K. (2001) J. Cell Sci. 114 1829-1838 [DOI] [PubMed] [Google Scholar]
- 30.Vestweber, D., and Kemler, R. (1985) EMBO J. 4 3393-3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, J., Izvolsky, K. I., Qian, J., and Cardoso, W. V. (2005) J. Biol. Chem. 280 4834-4841 [DOI] [PubMed] [Google Scholar]
- 32.Lu, J., Qian, J., Izvolsky, K. I., and Cardoso, W. V. (2004) Dev. Biol. 273 418-435 [DOI] [PubMed] [Google Scholar]
- 33.Chandu, D., Huppert, S. S., and Kopan, R. (2006) J. Neurochem. 96 228-235 [DOI] [PubMed] [Google Scholar]
- 34.Marambaud, P., Shioi, J., Serban, G., Georgakopoulos, A., Sarner, S., Nagy, V., Baki, L., Wen, P., Efthimiopoulos, S., Shao, Z., Wisniewski, T., and Robakis, N. K. (2002) EMBO J. 21 1948-1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzog, E. L., Van Arnam, J., Hu, B., and Krause, D. S. (2006) Stem Cells 24 1986-1992 [DOI] [PubMed] [Google Scholar]
- 36.Ge, R., Rajeev, V., Ray, P., Lattime, E., Rittling, S., Medicherla, S., Protter, A., Murphy, A., Chakravarty, J., Dugar, S., Schreiner, G., Barnard, N., and Reiss, M. (2006) Clin. Cancer Res. 12 4315-4330 [DOI] [PubMed] [Google Scholar]
- 37.van Eyll, J. M., Pierreux, C. E., Lemaigre, F. P., and Rousseau, G. G. (2004) J. Cell Sci. 117 2077-2086 [DOI] [PubMed] [Google Scholar]
- 38.van Tuyl, M., Groenman, F., Kuliszewski, M., Ridsdale, R., Wang, J., Tibboel, D., and Post, M. (2005) Am. J. Physiol. 288 L672-L682 [DOI] [PubMed] [Google Scholar]
- 39.Post, L. C., Ternet, M., and Hogan, B. L. (2000) Mech. Dev. 98 95-98 [DOI] [PubMed] [Google Scholar]
- 40.Taichman, D. B., Loomes, K. M., Schachtner, S. K., Guttentag, S., Vu, C., Williams, P., Oakey, R. J., and Baldwin, H. S. (2002) Dev. Dyn. 225 166-175 [DOI] [PubMed] [Google Scholar]
- 41.Benedito, R., and Duarte, A. (2005) Gene Expr. Patterns 5 750-755 [DOI] [PubMed] [Google Scholar]
- 42.Chen, F., Desai, T. J., Qian, J., Niederreither, K., Lu, J., and Cardoso, W. V. (2007) Development 134 2969-2979 [DOI] [PubMed] [Google Scholar]
- 43.Cheng, H. T., Miner, J. H., Lin, M., Tansey, M. G., Roth, K., and Kopan, R. (2003) Development 130 5031-5042 [DOI] [PubMed] [Google Scholar]
- 44.Doerfler, P., Shearman, M. S., and Perlmutter, R. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9312-9317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geling, A., Steiner, H., Willem, M., Bally-Cuif, L., and Haass, C. (2002) EMBO Rep. 3 688-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, M. H., and Kopan, R. (2003) Dev. Biol. 263 343-359 [DOI] [PubMed] [Google Scholar]
- 47.Micchelli, C. A., Esler, W. P., Kimberly, W. T., Jack, C., Berezovska, O., Kornilova, A., Hyman, B. T., Perrimon, N., and Wolfe, M. S. (2003) FASEB J. 17 79-81 [DOI] [PubMed] [Google Scholar]
- 48.Weinmaster, G., and Kopan, R. (2006) Development 133 3277-3282 [DOI] [PubMed] [Google Scholar]
- 49.Que, J., Okubo, T., Goldenring, J. R., Nam, K. T., Kurotani, R., Morrisey, E. E., Taranova, O., Pevny, L. H., and Hogan, B. L. (2007) Development 134 2521-2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishii, Y., Rex, M., Scotting, P. J., and Yasugi, S. (1998) Dev. Dyn. 213 464-475 [DOI] [PubMed] [Google Scholar]
- 51.Kubbutat, M. H., Key, G., Duchrow, M., Schluter, C., Flad, H. D., and Gerdes, J. (1994) J. Clin. Pathol. 47 524-528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi, S., and Stanley, P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5234-5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitsiadis, T. A., Henrique, D., Thesleff, I., and Lendahl, U. (1997) Development 124 1473-1483 [DOI] [PubMed] [Google Scholar]
- 54.Hart, A., Papadopoulou, S., and Edlund, H. (2003) Dev. Dyn. 228 185-193 [DOI] [PubMed] [Google Scholar]
- 55.Medina, M., and Dotti, C. G. (2003) Cell. Signal. 15 829-841 [DOI] [PubMed] [Google Scholar]
- 56.Gassmann, M., Casagranda, F., Orioli, D., Simon, H., Lai, C., Klein, R., and Lemke, G. (1995) Nature 378 390-394 [DOI] [PubMed] [Google Scholar]
- 57.Heber, S., Herms, J., Gajic, V., Hainfellner, J., Aguzzi, A., Rulicke, T., von Kretzschmar, H., von Koch, C., Sisodia, S., Tremml, P., Lipp, H. P., Wolfer, D. P., and Muller, U. (2000) J. Neurosci. 20 7951-7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inagaki, M., Irie, K., Ishizaki, H., Tanaka-Okamoto, M., Morimoto, K., Inoue, E., Ohtsuka, T., Miyoshi, J., and Takai, Y. (2005) Development 132 1525-1537 [DOI] [PubMed] [Google Scholar]
- 59.Larue, L., Ohsugi, M., Hirchenhain, J., and Kemler, R. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8263-8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marschang, P., Brich, J., Weeber, E. J., Sweatt, J. D., Shelton, J. M., Richardson, J. A., Hammer, R. E., and Herz, J. (2004) Mol. Cell. Biol. 24 3782-3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmits, R., Filmus, J., Gerwin, N., Senaldi, G., Kiefer, F., Kundig, T., Wakeham, A., Shahinian, A., Catzavelos, C., Rak, J., Furlonger, C., Zakarian, A., Simard, J. J., Ohashi, P. S., Paige, C. J., Gutierrez-Ramos, J. C., and Mak, T. W. (1997) Blood 90 2217-2233 [PubMed] [Google Scholar]
- 62.Ikeya, T., and Hayashi, S. (1999) Development 126 4455-4463 [DOI] [PubMed] [Google Scholar]
- 63.Ghabrial, A. S., and Krasnow, M. A. (2006) Nature 441 746-749 [DOI] [PubMed] [Google Scholar]
- 64.Llimargas, M. (1999) Development 126 2355-2364 [DOI] [PubMed] [Google Scholar]
- 65.Bolos, V., Grego-Bessa, J., and de la Pompa, J. L. (2007) Endocr. Rev. 28 339-363 [DOI] [PubMed] [Google Scholar]
- 66.Brooker, R., Hozumi, K., and Lewis, J. (2006) Development 133 1277-1286 [DOI] [PubMed] [Google Scholar]
- 67.Hellstrom, M., Phng, L. K., Hofmann, J. J., Wallgard, E., Coultas, L., Lindblom, P., Alva, J., Nilsson, A. K., Karlsson, L., Gaiano, N., Yoon, K., Rossant, J., Iruela-Arispe, M. L., Kalen, M., Gerhardt, H., and Betsholtz, C. (2007) Nature 445 776-780 [DOI] [PubMed] [Google Scholar]
- 68.de Celis, J. F., and Bray, S. (1997) Development 124 3241-3251 [DOI] [PubMed] [Google Scholar]
- 69.Savill, N. J., and Sherratt, J. A. (2003) Dev. Biol. 258 141-153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.