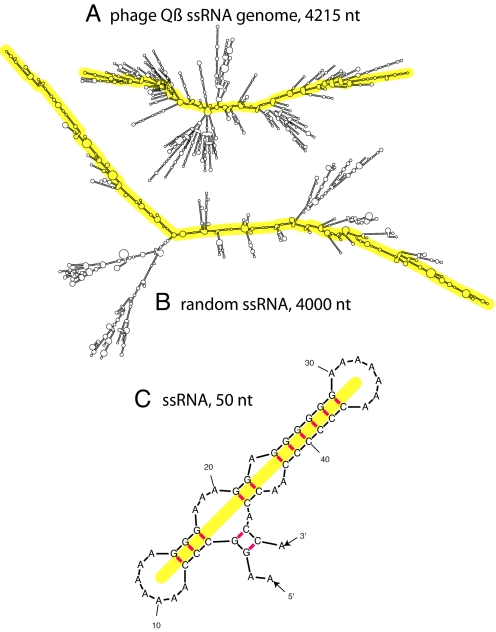
Fig. 1.
Predicted secondary structures of ssRNAs. (A) Enterobacteria phage Qß (in the Leviviridae family) ssRNA. (B) Randomly permuted ssRNA. Each is ≈4,000 nt in length and shown to the same scale. The MLDs of these structures are 221 and 368, respectively. (These are representative of their respective ensemble averages: The 〈MLD〉 of the phage Qß ssRNA is 240, and the 〈MLD〉 of 4,000-base random ssRNAs is 361.) The yellow overlays illustrate the paths associated with the MLDs (see text and the 50-nt example depicted in C). 〈MLD〉 values were calculated with RNAsubopt; figures were drawn with mfold.