Abstract
Sensory rhodopsin I (SRI) in Halobacterium salinarum acts as a receptor for single-quantum attractant and two-quantum repellent phototaxis, transmitting light stimuli via its bound transducer HtrI. Signal-inverting mutations in the SRI–HtrI complex reverse the single-quantum response from attractant to repellent. Fast intramolecular charge movements reported here reveal that the unphotolyzed SRI–HtrI complex exists in two conformational states, which differ by their connection of the retinylidene Schiff base in the SRI photoactive site to inner or outer half-channels. In single-quantum photochemical reactions, the conformer with the Schiff base connected to the cytoplasmic (CP) half-channel generates an attractant signal, whereas the conformer with the Schiff base connected to the extracellular (EC) half-channel generates a repellent signal. In the wild-type complex the conformer equilibrium is poised strongly in favor of that with CP-accessible Schiff base. Signal-inverting mutations shift the equilibrium in favor of the EC-accessible Schiff base form, and suppressor mutations shift the equilibrium back toward the CP-accessible Schiff base form, restoring the wild-type phenotype. Our data show that the sign of the behavioral response directly correlates with the state of the connectivity switch, not with the direction of proton movements or changes in acceptor pKa. These findings identify a shared fundamental process in the mechanisms of transport and signaling by the rhodopsin family. Furthermore, the effects of mutations in the HtrI subunit of the complex on SRI Schiff base connectivity indicate that the two proteins are tightly coupled to form a single unit that undergoes a concerted conformational transition.
Keywords: intramolecular charge movements, phototaxis, proton transport, receptor activation, receptor–transducer signal relay
Genomic and metagenomic studies have revealed >4,800 members of the rhodopsin family spread throughout the microbial world in both prokaryotic and eukaryotic microorganisms (1–4). From their primary structures, most of the microbial rhodopsins appear to be light-driven proton pumps (2). Phylogenetic analysis argues for multiple independent occurrences of duplication of a proton-pumping rhodopsin gene and its evolutionary modification into a light-sensing receptor that transmits signals to signal transduction machinery already present in the cell (3). This hypothesis is strongly supported by biochemical studies of sensory rhodopsins that reveal a variety of unrelated signal transduction pathways to which they are coupled (5). The best-characterized example of the evolutionary conversion from pump to sensor are the proton-transporter bacteriorhodopsin [BR (6–8)] and the two phototaxis receptors sensory rhodopsins I and II (SRI and SRII) in the archaeon Halobacterium salinarum (5, 9–12). SRI and SRII are photoactive subunits in membrane-embedded signaling complexes containing transducers (HtrI and HtrII, respectively) that are homologous to chemotaxis receptors in the same cell, and the phototaxis transducers transmit signals through the chemotaxis histidine kinase controlling the cell's motility.
Nearly 4 decades of study since the discovery of BR (13) have made light-driven proton transport by BR one of the best-understood functions of any membrane protein in terms of atomic and chemical mechanism (6–8, 14, 15). Two findings have revealed that BR transport and SR signaling mechanisms are closely related. First was the observation that the SRI subunit expressed in cells without HtrI carries out light-driven proton transport, although at a slow rate in comparison with BR (16, 17). In complex with HtrI, however, proton transport activity by SRI is suppressed (18, 19). A similar but weaker residual proton transport activity was later found in transducer-free SRII, similarly blocked by binding of its transducer HtrII (20, 21). Therefore, the essential components of BR transport are present at least to some extent in the isolated SR proteins but may not be present and in any case do not result in transport in their natural states in complex with their transducers.
The second finding is that BR genetically engineered to bind to HtrII and to contain a photoactive site steric trigger known to be essential for SRII signal relay to HtrII enabled BR to signal to HtrII and mediate phototaxis responses (22). This study showed that essential components of the SRII signaling mechanism, other than the transducer interaction surface and the steric trigger, are evidently already present in BR.
Although only ≈25% identical to BR in primary structure, the SRs share with BR a seven-transmembrane helix structure that forms an interior pocket for the chromophore retinal, which is attached in a protonated Schiff base linkage to the ε-amino group of a lysyl residue. The protonated Schiff base plays a central role in proton translocation by BR, deprotonating in the first half of the pumping cycle by transferring its proton to an Asp residue in the extracellular half-channel and reprotonating in the second half of the cycle from an Asp residue in the cytoplasmic channel. The key step in this proton pumping process is the “Schiff base connectivity switch” (7, 23–25), the photoactive site structural change that occurs in the pumping cycle in BR and changes the accessibility of the retinylidene Schiff base nitrogen from the extracellular to the cytoplasmic channel. The connectivity switch creates the alternate side access essential for vectorial transmembrane proton movement. The residual proton pumping activity by transducer-free SRs implies that the Schiff base connectivity switch is at least partially functional in SR proteins (26). Two critical questions are as follows: (i) Does this switch occur also in SRs in their signaling complexes, which do not carry out proton transport? And (ii) if the Schiff base connectivity switch occurs in the complexes, is it functionally important in signal relay to the transducers? Here we answer these questions by measurements of Schiff base proton displacements in SRI–HtrI complexes with signal-inverting mutations in the SRI and HtrI subunits and their suppressors, which reveal a definitive role of the Schiff base connectivity switch in signaling.
Results
Transducer-Free SRI.
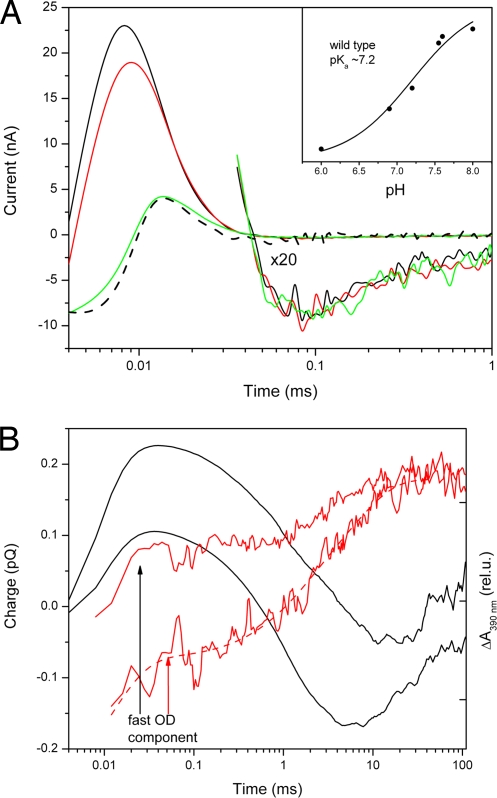
A 532-nm laser flash generates two resolved oppositely directed currents in free SRI in Escherichia coli cells at our time resolution of 4 μs (Fig. 1A). A fast current with decay time in the range of 10 μs corresponds to outwardly directed positive charge movement (positive current) and strongly dominates. The positive current is followed by an orders-of-magnitude smaller and slower cytoplasmically directed current. Acidification dramatically reduces the positive current with pKa ≈ 7.2 (Fig. 1A Inset), whereas the slow negative current is pH-independent in the range examined. Mutation to Asn of Asp-76, the homolog to the Schiff base proton acceptor in BR and SRII, previously shown to be a Schiff base proton acceptor in purified SRI at alkaline pH (27), eliminates the fast positive photocurrent (Fig. 1A).
Fig. 1.
Laser flash-induced charge movements and absorption changes in free SRI. (A) Photocurrents in wild-type SRI (solid lines) at pH 7.2 (black), 6.9 (red), and 5.4 (green) and in the SRI_D76N mutant at pH 8.0 (dashed black line). Slow region of the signals are duplicated in 20× magnification. (Inset) The pH dependence of maximum current in the wild-type SRI. (B) Charge movement (integral of photocurrent over time) (left axis, black lines) and M intermediate accumulation (absorption changes at 390 nm) (right axis, red lines) in wild-type SRI at pH 7.1 (upper curves) and 6.3 (lower curves). Absorption changes were normalized at their maximum value. Arrows show an increase in the relative amplitude of the fast component of M accumulation corresponding to an increase in the fast outwardly directed charge movement.
Integration of the current over time yields the overall charge displacement (28). The positive (outwardly directed toward the extracellular side) charge movement correlates with the fast component of Schiff base deprotonation as measured by accumulation of the blue-shifted M (also called S373) photointermediate in the same sample. The negative (cytoplasmically directed) displacement of positive charge correlates with the slow component of Schiff base deprotonation. Upon alkalization the amplitude of the fast positive charge movement increases in parallel with the fast component of M formation and becomes dominant at neutral and high pH (Fig. 1B).
These data show that free SRI in intact E. coli cells behaves spectroscopically the same as in natural H. salinarum membranes, where M formation with very similar values for the fast (≈10 μs) and slow (≤10 ms) components was found (29). Additionally, we show that the two phases of M formation result from two oppositely directed Schiff base deprotonation reactions: one toward Asp-76 located in the outer half-channel and a second toward an unknown acceptor in the cytoplasmic half-channel. The pH-dependent changes in the relative amplitudes of fast and slow M accumulation correlate with the amplitudes of positive and negative charge displacements, proving that the latter reflect alternative Schiff base deprotonation reactions, and not a successive charge movement (e.g., reprotonation of the Schiff base from Asp-76).
SRI–HtrI Complex.
Charge movements in SRI–HtrI complexes dramatically differ from those in free SRI (Fig. 2A). The fast outwardly directed charge movement is greatly reduced in amplitude. Instead biphasic charge movement toward the cytoplasmic side of the molecule that peaks at several milliseconds dominates.
Fig. 2.
Laser flash-induced charge movements and absorption changes in wild-type SRI–HtrI complex. (A) Charge movements in SRI–HtrI complex (red line) and in free SRI (black line) are compared. (B) Effect of deuteration on the kinetics of charge movements (left axis, solid lines without fitting curves) and M intermediate accumulation (absorption changes at 390 nm) (right axis with dashed fitting curves) in SRI–HtrI complex. Black lines, H2O; red lines, D2O.
Deuteration of the sample slows the positive and negative charge displacements, confirming that they reflect proton movements. As in free SRI, the small outwardly directed proton transfer correlates with the fast component of M intermediate accumulation and the cytoplasmically directed proton transfer with the slow component in both H2O and D2O (Fig. 2B).
Charge Movement in Behavioral Mutants of SRI–HtrI.
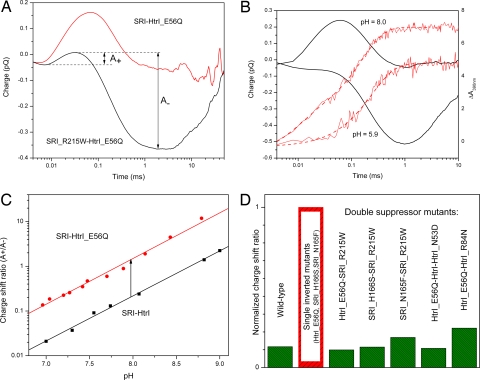
Orange light elicits an attractant motility response [light-induced transient inhibition of reversals in swimming direction (5, 9–12)] in H. salinarum containing wild type SRI–HtrI complexes. A number of mutations in SRI and HtrI invert the sign of the single-quantum motility response from attractant to repellent, and second-site suppressors restore the motility response to the wild-type attractant phenotype (Fig. 3) (30). The inverting mutations strongly activate the outwardly directed Schiff base deprotonation reaction and suppress the cytoplasmically directed proton transfer. Suppressor mutations, which restore wild-type motility behavior to cells carrying inverting mutations (30), also restore the wild-type pattern of proton movement (Fig. 4A).
Fig. 3.
Structural schematic of SRI–HtrI. Shown are locations of amino acid residues that when mutated lead to inverted (marked in red) or recovered (suppressor mutations, marked in green) phototaxis responses. The interaction of SRI and HtrI is schematically presented based on analogy with the SRII–HtrII (40) and HAMP domain (50) atomic structures.
Fig. 4.
Laser flash-induced charge movements in inverted and suppressor mutants. (A) Kinetics of charge movements in the inverted (SRI–HtrI_E56Q) (upper red line) and suppressor (SRI_R215W-HtrI_E56Q) (lower black line) mutants. A+ and A−, amplitudes of outwardly and inwardly directed proton movements, respectively. (B) Correlation between charge movements and M accumulation in double mutant SR_R215W-HtrI_E56Q. Shown are charge movements (left scale, black lines), and M intermediate accumulation (absorption change at 390 nm) (right scale, red lines) at pH 5.9 (lower curves) and pH 8.0 (upper curves). (C) pH dependence of the ratio of outwardly and inwardly directed proton movements (A+/A−) in wild-type SRI–HtrI (black symbols and line) and the inverted SRI–HtrI_E56Q mutant (red symbols and line). (D) Effect of various mutations in receptor and transducer subunits of the SRI–HtrI complex on the ratio of fast outwardly directed charge movement to slower inwardly directed charge movement (A+/A−). The ratio values for double suppressor mutants were normalized to corresponding single inverted mutants. The wild-type ratio was normalized to the inverted SRI–HtrI_E56Q mutant.
In all tested mutants, as well as in the wild-type complex, the relative amplitudes of oppositely directed charge movements change with pH in parallel with the relative amplitudes of fast and slow M accumulation, confirming that they derive from competitive and not sequential proton transfer reactions (see Fig. 4B as an example).
The ratio of positive charge shift (A+) to negative charge shift (A−) (Fig. 4A) provides a measure of the relative amounts of SRI–HtrI complex molecules with outwardly and cytoplasmically directed Schiff base deprotonation, respectively. This ratio is pH-dependent, and this dependence is roughly linear on a semilogarithmic scale (Fig. 4C). We used the vertical shift of the curves to characterize 3 inverting mutations individually (HtrI_E56Q, SRI_H166S, and SRI_N165F) and in combination with the suppressor mutations SRI_215W, HtrI_N53D, and HtrI_R84N.
Each inverting mutation tested increases the ratio of outward to inward Schiff base proton transfer by ≈3-, 8-, and 30-fold for N165F, E56Q, and H166S, respectively. Each of the 3 suppressor mutations, so named because they return the cells with inverting mutations to near wild-type motility behavior, also decreases the ratio (toward wild-type values) from 5- to 10-fold (Fig. 4D).
Schiff Base Connectivity Is to Opposite Sides of the Protein in the Two Opposite Signaling Forms of SRI–HtrI.
In the above measurements attractant and repellent motility responses are shown to perfectly correlate with inwardly directed and outwardly directed Schiff base deprotonation, respectively. One possible explanation is that the proton movement to the inner or outer half-channel itself triggers the behavioral response of corresponding sign. This possibility is ruled out because we can make mutations that invert the wild-type motility behavior without appearance of outwardly directed proton movement. Specifically, neutralizing Asp-76 by mutation to Asn does not alter the wild-type attractant behavior of H. salinarum (31). No outwardly directed proton transfer is observed in the D76N mutant (Fig. 5), which corresponds to the predominantly inwardly directed Schiff base deprotonation characteristic of wild-type SRI–HtrI (see above). We examined the motility behavior of the double mutant SRI_D76N-HtrI_E56Q and observe that the additional E56Q mutation inverts the behavioral response of SRI_D76N from attractant to repellent without the appearance of any positive current in contrast to the effect of this mutation in the wild type (Fig. 5). Therefore, the proton movement per se to the inner or outer half-channel does not determine the sign of the behavioral response and, moreover, is evidently not significantly involved in the sensory signaling process. The direction of proton movement is a passive consequence of the attractant and repellent signaling state conformations and is a useful measure of their relative amounts.
Fig. 5.
Charge movements in D76N mutants. Shown are phenotypically wild-type SRI_D76N-HtrI single mutant (green lines), inverted SRI–HtrI_E56Q single mutant (red lines), and inverted SRI_D76N-HtrI_E56Q double mutant (black lines). Arrow shows the decrease in the amplitude of inwardly directed proton movement.
There are two possibilities for the altered direction of Schiff base deprotonation in the behavioral mutants: (i) inner and outer acceptors compete for the Schiff base proton, and their relative pKa values are changed by the mutations; or (ii) the relative sizes of two subpopulations of SRI–HtrI complex with opposite Schiff base connectivity are changed by the mutations. The data in Fig. 5 exclude that the extent of inwardly directed proton release is simply controlled by the pKa of Asp-76, which competes for acceptance of the proton. Despite the absence of the Asp-76 acceptor in the outer half-channel and the corresponding absence of positive photocurrent, the amplitude of inwardly directed proton movement is greatly decreased in the signal-inverted double mutant (Fig. 5). Evidently the relative sizes of subpopulations of the complex poised to deprotonate the Schiff base to the outer or inner half-channels, regardless of whether an acceptor is available, i.e., the Schiff base connectivity switch, determine the sign of the phototaxis signal. The conformer with inwardly connected Schiff base mediates the attractant response, whereas the conformer with opposite connectivity mediates the opposite repellent response.
Discussion
The findings above can be summarized in Fig. 6. In the dark the SRI–HtrI complex exists in two conformational states, which differ by their connectivity of the Schiff base to the inner or outer half-channels. In single-quantum photochemical reactions, the conformer with the Schiff base connected to the cytoplasmic (CP) half-channel generates an attractant signal, whereas the conformer with the Schiff base connected to the extracellular (EC) half-channel generates a repellent signal. The opposite signals from the two conformers are integrated in the downstream transduction pathways leading to one or the other sign of the behavioral response. In the wild-type complex the equilibrium between the conformers is poised strongly in favor of that with CP-accessible Schiff base. Signal-inverting mutations shift the equilibrium between the two conformers in favor of the EC-accessible Schiff base form, thereby inverting the motility response, and second-site suppressor mutations shift the equilibrium back toward the CP-accessible Schiff base form, restoring the wild-type response phenotype.
Fig. 6.
Schematic representation. Cartoon depicts light signal transduction by two conformers of the SRI–HtrI complex with opposite Schiff base connectivity assuming switch-coupled channel conformations (see text).
The data show that the sign of the behavioral response directly correlates with the state of the connectivity switch, not with the direction of Schiff base proton movements or changes in acceptor pKa. When the Asp-76 Schiff base counterion is present, the direction of proton release from the Schiff base serves as an indicative measure of the relative sizes of subpopulation of SRI–HtrI complex with opposite Schiff base connectivity. However, proton movement in the opposite direction is a consequence rather than a cause of signal-inverting mutations.
We show here that the inverted repellent-signaling mutant conformer of SRI–HtrI has the same EC-oriented Schiff base connectivity as unphotolyzed BR. Conversely, the wild-type attractant-signaling SRI–HtrI conformer resembles in this respect the M2 or N intermediates of BR with opposite connectivity (32, 33). Therefore, our finding identifies a shared fundamental process in the mechanisms of transport and signaling by microbial rhodopsins. The Schiff base connectivity switch is not only preserved in SRI, but also plays a key role in its signaling function. Interestingly, in the second haloarchaeal sensory rhodopsin complex (SRII–HtrII), which mediates repellent responses, Schiff base connectivity is also directed toward the EC as in the repellent conformer of SRI and only outwardly directed proton transfer is observed (34).
The hypothesis of two conformational states of the SRI–HtrI with opposite signaling signs was proposed based on behavioral responses of a number of signaling mutants (30) and spin-label detection of conformational shifts caused by the mutations in the dark (35). Here we experimentally confirmed this hypothesis and identify the Schiff base connectivity as a key structural difference in the two conformers.
Mutations in HtrI as well as in SRI invert Schiff base connectivity in the photoactive site of SRI and, correspondingly, the sign of the behavioral response. Furthermore, the effect of inverting mutations in either subunit can be recovered by second-site mutations in this subunit or in its counterpart. Even distant mutations in the membrane-proximal parts of the transducer HAMP domain control Schiff base connectivity in the photoactive site of SRI. Our interpretation is that the SRI subunit and adjacent parts of HtrI, including the membrane-proximal portion of the HAMP domain, function as a single unit that undergoes concerted conformational transitions.
The chemical nature of the connectivity switch in the retinylidene photoactive site remains largely unknown. In BR this switch takes place during transition between two conformational states with closed and open cytoplasmic half-channels (M1 and M2 states) (14, 36, 37). Channel opening in the M2 state is achieved by an outward tilting of helix F accompanied by a smaller helix G displacement. Changes in helix position themselves are not sufficient to switch the connectivity of the Schiff base, because a triple mutant of BR with an open cytoplasmic half-channel-like conformation is still capable of transporting protons in the normal outward direction in a single-quantum reaction cycle (38, 39). Nevertheless, such a conformational transition occurs and is involved in normally functioning BR. In the SRII–HtrII complex, helices F and G are in extensive contact with the transmembrane helices of HtrII (40), and based on several lines of indirect evidence outward tilting of helix F similar to that in BR has been suggested to be involved in transmitting the SRII signal to its transducer (41, 42).
Because in the dominating conformer of the wild-type SRI–HtrI light-induced Schiff base deprotonation is toward the cell interior, the unphotolyzed SRI protein may resemble the conformation of the M2 intermediate of BR, in which the Schiff base is connected to the cytoplasmic side. Assuming analogous conformational rearrangements in sensory rhodopsins and BR one can envision that in SRI single-quantum absorption produces a cytoplasmic-side closed conformation, i.e., with untilted F helix and with the Schiff base connected to the outer half-channel, as in the unphotolyzed states of BR and SRII. With this assumption, this closed channel conformer would evidently produce an attractant signal. Such a conformational change would be opposite to that produced in the SRII photocycle, which by analogy with BR is expected to produce a transient cytoplasmic-side open conformer that mediates the opposite (repellent) responses (9). If cytoplasmic channel opening and closing contribute to the signaling, then the 2-quantum photoreaction of the SRI–HtrI complex, which generates a repellent-signaling state, would be expected to produce the open cytoplasmic-side channel conformation.
Some microbial rhodopsins exhibit two retinylidene chromophore isomeric configurations, all trans and 13-cis, in the population of molecules in the dark state, e.g., BR (43, 44) and Anabaena sensory rhodopsin (ASR) (45). Some BR mutants are thermally stable in a conformation with the Schiff base connected to the inner half-channel achieved by a 13-cis, 15-anti configuration of the retinal chromophore (32). By analogy the inversed connectivity in wild-type SRI–HtrI with respect to free SRI or the inverted mutants could be also achieved by an initially isomerized chromophore state. However, the dark state isomer configuration of the retinylidene chromophore in the wild-type SRI–HtrI complex is all trans, with no detectable 13-cis isomer present (46). Like the all trans isomeric forms of BR and ASR (47), upon formation of M, SRI undergoes all trans to 13-cis photoisomerization in both free and complex form (48). FTIR spectroscopy does not show significant differences between the wild-type complex and inverted SRI–HtrI_E56Q mutant complex in reconstituted liposomes (O.A.S., J.S., B.J.P., J.L.S., and Hideki Kandori, unpublished data). Thus, direct correlation between the isomeric configuration of the chromophore and the Schiff base connectivity in SRI is unlikely, although we cannot fully exclude it in intact cells.
Materials and Methods
Plasmid Construction and Expression.
HtrI-free SRI and SRI–HtrI1–147 fusion (truncated at position 147 in HtrI) were cloned into pET21d modified by using NcoI and BamHI restriction cleavage sites (49). Residue mutations were introduced by the two-step mega-primer PCR method with Pfu turbo polymerase as described earlier (49).
Photoinduced Current Measurements.
Intramolecular charge movements were measured in suspension of E. coli cells expressing desired protein by the method described earlier (34). A suspension of E. coli cells was flashed by Nd:YAG Surelite-I laser (532 nm, 6-ns pulse; Continuum) along the line between two platinum electrodes. A macroscopic electrical current in the cuvette appeared due to asymmetric absorption of light in each bacterial cell. The electrode remote from the light source was fed into a low-noise current amplifier 428 (Keithley) with 2-μs rise time. The signals were digitized and stored by using the DIGI-DATA 1325A and pCLAMP 9.0 program (both from Axon Instruments). Twenty to 150 signals with maximum sampling rate of 2 μs per point were averaged. If not otherwise indicated, the measuring buffer contained 5 mM Tris·HCl, 1.5 mM NaCl, 0.15 mM CaCl2 and 0.15 mM MgSO4 (pH 7.6).
Absorption and Laser Flash Photolysis.
Flash-induced absorption changes were acquired in parallel with a laboratory-constructed cross-beam laser flash-photolysis system under conditions identical to those of the current measurements as described (22).
Other Software.
Protein sequence analysis used DNAStar V6.0 (DNASTAR), and protein modeling used Accelrys DS Visualizer 1.7 (Accelrys).
Acknowledgments.
We thank Elena Spudich for valuable discussions. This work was supported by National Institutes of Health Grant R37GM27750, Department of Energy Grant DE-FG02-07ER15867, and an endowed chair from the Robert A. Welch Foundation (to J.L.S.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Ruiz-Gonzalez MX, Marin I. New insights into the evolutionary history of type 1 rhodopsins. J Mol Evol. 2004;58:348–358. doi: 10.1007/s00239-003-2557-8. [DOI] [PubMed] [Google Scholar]
- 2.Spudich JL, Jung KH. In: Handbook of Photosensory Receptors. Briggs WR, Spudich JL, editors. Weinheim, Germany: Wiley VCH; 2005. pp. 1–21. [Google Scholar]
- 3.Sharma AK, Spudich JL, Doolittle WF. Microbial rhodopsins: Functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spudich JL. The multitalented microbial sensory rhodopsins. Trends Microbiol. 2006;14:480–487. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Stoeckenius W. Bacterial rhodopsins: Evolution of a mechanistic model for the ion pumps. Protein Sci. 1999;8:447–459. doi: 10.1110/ps.8.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haupts U, Tittor J, Oesterhelt D. Closing in on bacteriorhodopsin: Progress in understanding the molecule. Annu Rev Biophys Biomol Struct. 1999;28:367–399. doi: 10.1146/annurev.biophys.28.1.367. [DOI] [PubMed] [Google Scholar]
- 8.Lanyi JK, Luecke H. Bacteriorhodopsin Curr Opin Struct Biol. 2001;11:415–419. doi: 10.1016/s0959-440x(00)00226-8. [DOI] [PubMed] [Google Scholar]
- 9.Hoff WD, Jung K-H, Spudich JL. Molecular machanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 10.Kamo N, Shimono K, Iwamoto M, Sudo Y. Photochemistry and photoinduced proton-transfer by pharaonis phoborhodopsin. Biochemistry (Moscow) 2001;66:1277–1282. doi: 10.1023/a:1013187403599. [DOI] [PubMed] [Google Scholar]
- 11.Klare JP, Chizhov I, Engelhard M. Microbial rhodopsins: Scaffolds for ion pumps, channels, and sensors. Results Probl Cell Differ. 2008;45:73–122. doi: 10.1007/400_2007_041. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki J, Spudich JL. Signal transfer in haloarchaeal sensory rhodopsin- transducer complexes. Photochem Photobiol. 2008;84:863–868. doi: 10.1111/j.1751-1097.2008.00314.x. [DOI] [PubMed] [Google Scholar]
- 13.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 14.Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 15.Kandori H. Hydration switch model for the proton transfer in the Schiff base region of bacteriorhodopsin. Biochim Biophys Acta. 2004;1658:72–79. doi: 10.1016/j.bbabio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Bogomolni RA, et al. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc Natl Acad Sci USA. 1994;91:10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haupts U, Bamberg E, Oesterhelt D. Different modes of proton translocation by sensory rhodopsin I. EMBO J. 1996;15:1834–1841. [PMC free article] [PubMed] [Google Scholar]
- 18.Olson KD, Spudich JL. Removal of transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993;65:2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spudich JL. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994;79:747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 20.Sudo Y, et al. Photo-induced proton transport of pharaonis phoborhodopsin (sensory rhodopsin II) is ceased by association with the transducer. Biophys J. 2001;80:916–922. doi: 10.1016/S0006-3495(01)76070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmies G, et al. Electrophysiological characterization of specific interactions between bacterial sensory rhodopsins and their transducers. Proc Natl Acad Sci USA. 2001;98:1555–1559. doi: 10.1073/pnas.031562298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudo Y, Spudich JL. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc Natl Acad Sci USA. 2006;103:16129–16134. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kataoka M, et al. Energy coupling in an ion pump. The reprotonation switch of bacteriorhodopsin. J Mol Biol. 1994;243:621–638. doi: 10.1016/0022-2836(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 24.Brown LS, Dioumaev AK, Needleman R, Lanyi JK. Connectivity of the retinal Schiff base to Asp85 and Asp96 during the bacteriorhodopsin photocycle: The local-access model. Biophys J. 1998;75:1455–1465. doi: 10.1016/S0006-3495(98)74064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 26.Spudich JL, Lanyi JK. Shuttling between two protein conformations: The common mechanism for sensory transduction and ion transport. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 27.Rath P, et al. Asp76 is the Schiff base counterion and proton acceptor in the proton-translocating form of sensory rhodopsin I. Biochemistry. 1996;35:6690–6696. doi: 10.1021/bi9600355. [DOI] [PubMed] [Google Scholar]
- 28.Der A, et al. Interpretation of the spatial charge displacements in bacteriorhodopsin in terms of structural changes during the photocycle. Proc Natl Acad Sci USA. 1999;96:2776–2781. doi: 10.1073/pnas.96.6.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung KH, Spudich EN, Dag P, Spudich JL. Transducer-binding and transducer-mutations modulate photoactive-site deprotonation in sensory rhodopsin I. Biochemistry. 1999;38:13270–13274. doi: 10.1021/bi991180w. [DOI] [PubMed] [Google Scholar]
- 30.Jung K-H, Spudich JL. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: Insights into the color-sensing mechanism. J Bacteriol. 1998;180:2033–2042. doi: 10.1128/jb.180.8.2033-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rath P, Olson KD, Spudich JL, Rothschild KJ. The Schiff base counterion of bacteriorhodopsin is protonated in sensory rhodopsin I: Spectroscopic and functional characterization of the mutated proteins D76N and D76A. Biochemistry. 1994;33:5600–5606. doi: 10.1021/bi00184a032. [DOI] [PubMed] [Google Scholar]
- 32.Dioumaev AK, Brown LS, Needleman R, Lanyi JK. Partitioning of free energy gain between the photoisomerized retinal and the protein in bacteriorhodopsin. Biochemistry. 1998;37:9889–9893. doi: 10.1021/bi980934o. [DOI] [PubMed] [Google Scholar]
- 33.Lanyi JK, Schobert B. Local-global conformational coupling in a heptahelical membrane protein: Transport mechanism from crystal structures of the nine states in the bacteriorhodopsin photocycle. Biochemistry. 2004;43:3–8. doi: 10.1021/bi035843s. [DOI] [PubMed] [Google Scholar]
- 34.Sineshchekov OA, Spudich JL. Light-induced intramolecular charge movements in microbial rhodopsins in intact E. coli cells. Photochem Photobiol Sci. 2004;3:548–554. doi: 10.1039/b316207a. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki J, et al. Different dark conformations function in color-sensitive photosignaling by the sensory rhodopsin I-HtrI complex. Biophys J. 2007;92:4045–4053. doi: 10.1529/biophysj.106.101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorgeirsson TE, et al. Transient channel-opening in bacteriorhodopsin: An EPR study. J Mol Biol. 1997;273:951–957. doi: 10.1006/jmbi.1997.1362. [DOI] [PubMed] [Google Scholar]
- 37.Subramaniam S, et al. Protein conformational changes in the bacteriorhodopsin photocycle. J Mol Biol. 1999;287:145–161. doi: 10.1006/jmbi.1999.2589. [DOI] [PubMed] [Google Scholar]
- 38.Subramaniam S, Hirai T, Henderson R. From structure to mechanism: Electron crystallographic studies of bacteriorhodopsin. Philos Transact A Math Phys Eng Sci. 2002;360:859–874. doi: 10.1098/rsta.2001.0971. [DOI] [PubMed] [Google Scholar]
- 39.Tittor J, et al. Proton translocation by bacteriorhodopsin in the absence of substantial conformational changes. J Mol Biol. 2002;319:555–565. doi: 10.1016/S0022-2836(02)00307-8. [DOI] [PubMed] [Google Scholar]
- 40.Gordeliy VI, et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419:484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 41.Spudich JL. Variations on a molecular switch: Transport and sensory signalling by archaeal rhodopsins. Mol Microbiol. 1998;28:1051–1058. doi: 10.1046/j.1365-2958.1998.00859.x. [DOI] [PubMed] [Google Scholar]
- 42.Klare JP, et al. The archaeal sensory rhodopsin II/transducer complex: A model for transmembrane signal transfer. FEBS Lett. 2004;564:219–224. doi: 10.1016/S0014-5793(04)00193-0. [DOI] [PubMed] [Google Scholar]
- 43.Balashov SP, et al. Evidence that aspartate-85 has a higher pK(a) in all-trans than in 13-cis bacteriorhodopsin. Biophys J. 1996;71:1973–1984. doi: 10.1016/S0006-3495(96)79395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dencher NA, Kohl KD, Heyn MP. Photochemical cycle and light-dark adaptation of monomeric and aggregated bacteriorhodopsin in various lipid environments. Biochemistry. 1983;22:1323–1334. doi: 10.1021/bi00275a002. [DOI] [PubMed] [Google Scholar]
- 45.Vogeley L, et al. Anabaena sensory rhodopsin: A photochromic color sensor at 2.0 A. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan B, Nakanishi K, Spudich JL. Mechanism of activation of sensory rhodopsin I: Evidence for a steric trigger. Proc Natl Acad Sci USA. 1991;88:9412–9416. doi: 10.1073/pnas.88.21.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sineshchekov OA, Trivedi VD, Sasaki J, Spudich JL. Photochromicity of Anabaena sensory rhodopsin, an atypical microbial receptor with a cis-retinal light-adapted form. J Biol Chem. 2005;280:14663–14668. doi: 10.1074/jbc.M501416200. [DOI] [PubMed] [Google Scholar]
- 48.Haupts U, Eisfeld W, Stockburger M, Oesterhelt D. Sensory rhodopsin I photocycle intermediate SRI380 contains 13-cis retinal bound via an unprotonated Schiff base. FEBS Lett. 1994;356:25–29. doi: 10.1016/0014-5793(94)01226-1. [DOI] [PubMed] [Google Scholar]
- 49.Furutani Y, et al. Structural changes of sensory rhodopsin I and its transducer protein are dependent on the protonated state of Asp76. Biochemistry. 2008;47:2875–2883. doi: 10.1021/bi702050c. [DOI] [PubMed] [Google Scholar]
- 50.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]