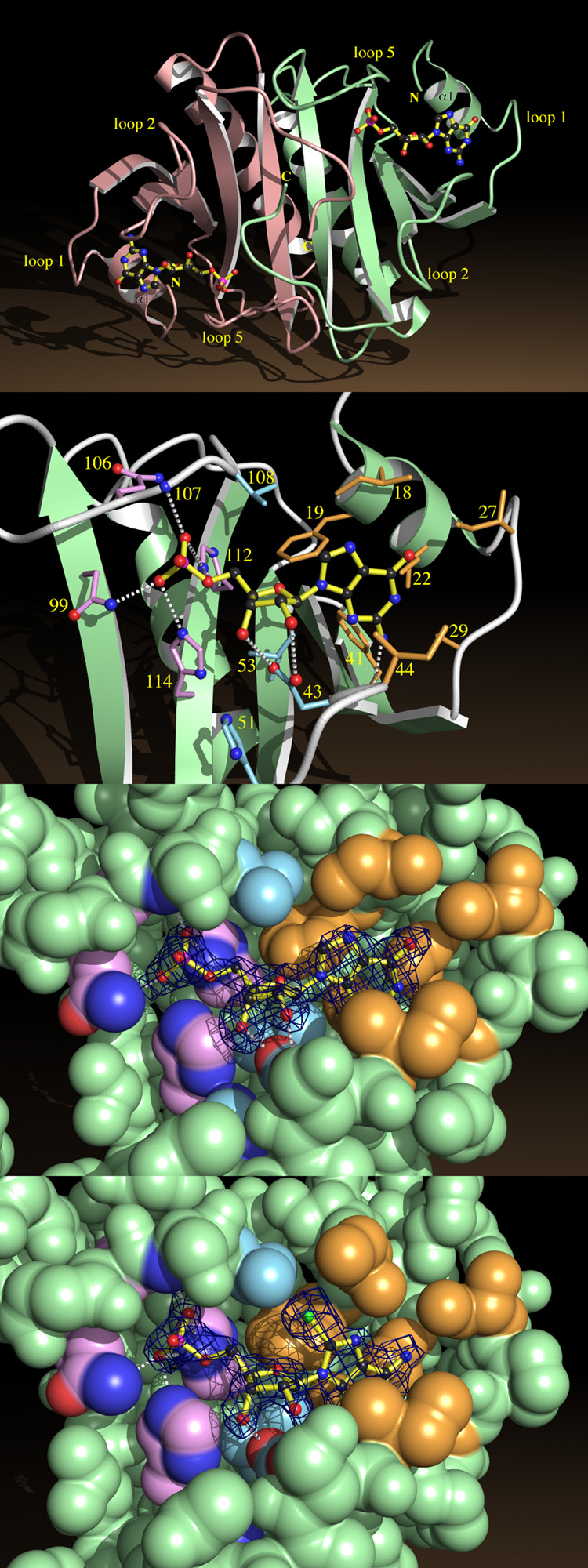
Fig. 4.
Nucleotide Binding by HINT.
a, HINT-GMP dimer shown from above the β-sheet, indicating helices α1 and loops 1, 2 and 5.
b, A close-up of GMP recognition by HINT including nonhydrogen atoms of GMP and side chains from fifteen conserved residues. Side chains surrounding the guanine base are beige, those surrounding ribose are blue, and those surrounding phosphate are purple. Potential hydrogen bonds between GMP and protein are indicated.
c, Coordinates corresponding to GMP in the HINT-GMP structure were removed and the protein model was subjected to simulated annealing at 3000 °C50. A difference electron density map was calculated with coefficients (Fobs - Fcalc) and contoured at 2.4 σ over the omitted GMP coordinates. Agreement between the map and model of GMP demonstrates that phases calculated from the protein model are robust and that GMP binds in this manner. The nucleotide-binding site is represented by its van der Waals surface, color coded as above.
d, A simulated annealing omit map prepared, as above, for HINT-8-Br-AMP.