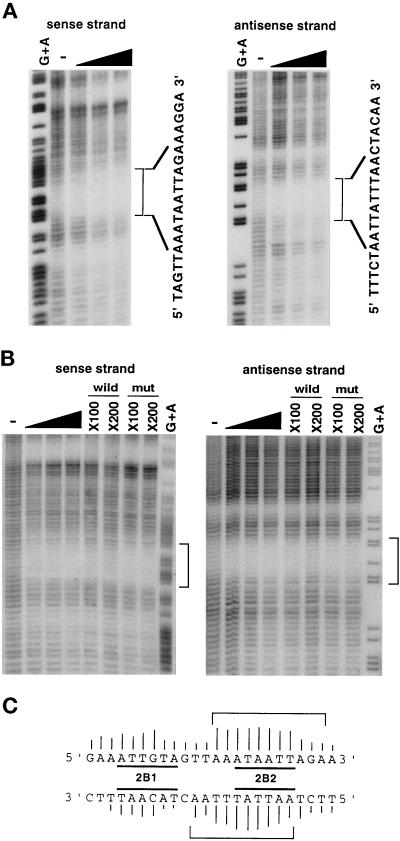
Figure 1.
Chemical footprint analysis for the Orpheus-binding site of ISE2. (A) The footprint patterns on the sense and antisense strands of ISE2 with the partially purified nuclear extract of IML12–4. Fifteen, 30, and 45 U of the Orpheus-binding activity were used in this assay, as indicated by black wedges. The specific footprints of Orpheus are indicated by square brackets with the nucleotide sequences of ISE2. Lanes of G+A and − represent the sequence ladders produced by GA-specific Maxam-Gilbert sequencing and hydroxyl radical cleavage reactions for the intact DNA fragments, respectively. (B) The similar footprint patterns were observed with the partially purified nuclear extract of HeLa-S3, as indicated by square brackets. The footprints of Orpheus were inhibited by the addition of indicated molar excess of the wild-type oligonucleotides, but not by that of mutant-type oligonucleotides. (C) The Orpheus contact sites on the sequence of ISE2 are shown by square brackets. The height of bars on each nucleotide shows the relative strength of the contact between protein and the nucleotide. The 2B1 and 2B2 sites are previously determined as the negative regulatory elements by substitutive mutagenesis (Imai et al., 1994).