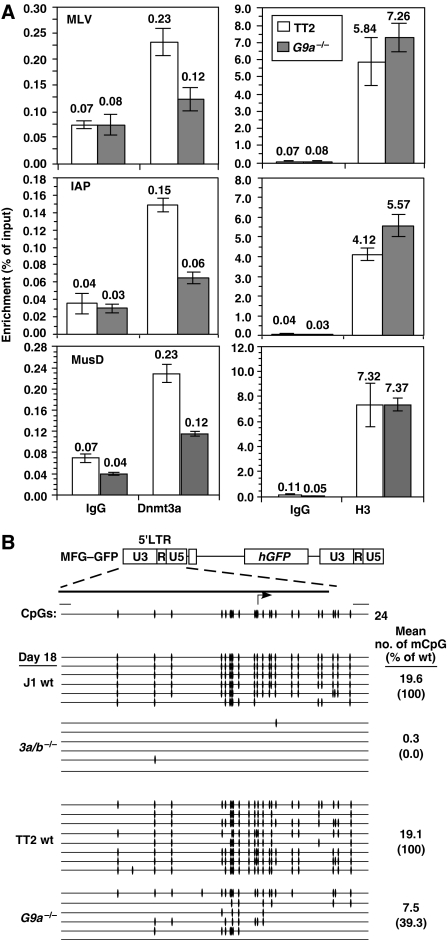
Figure 4.
G9a−/− ES cells show defects in recruitment of Dnmt3a to ERVs and de novo methylation of introduced retroviruses. (A) Formaldehyde-fixed chromatin was isolated from TT2 and G9a−/− lines and ChIP was conducted using nonspecific IgG or antisera raised against Dnmt3a or unmodified histone H3. Real-time PCR using primers specific for the LTR regions of MLV, IAP and MusD ERVs was carried out and values are presented as percentage of input precipitated (±s.d.) relative to the input in the representative experiment shown. A significant reduction in Dnmt3a enrichment in the G9a−/− line relative to the wt control is clearly apparent. (B) TT2 wt, G9a−/−, J1 wt and Dnmt3a/b−/− (3a/b−/−) lines were infected with the retroviral vector MFG–GFP and passaged in the absence of selection. Genomic DNA was isolated on day 18 post-infection and analysed by bisulphite genomic sequencing.