Abstract
The cycle of deacylation and reacylation of phospholipids plays a critical role in regulating availability of arachidonic acid for eicosanoid production. The major yeast lysophospholipid acyltransferase, Ale1p, is related to mammalian membrane-bound O-acyltransferase (MBOAT) proteins. We expressed four human MBOATs in yeast strains lacking Ale1p and studied their acyl-CoA and lysophospholipid specificities using novel mass spectrometry-based enzyme assays. MBOAT1 is a lysophosphatidylserine (lyso-PS) acyltransferase with preference for oleoyl-CoA. MBOAT2 also prefers oleoyl-CoA, using lysophosphatidic acid and lysophosphatidylethanolamine as acyl acceptors. MBOAT5 prefers lysophosphatidylcholine and lyso-PS to incorporate linoleoyl and arachidonoyl chains. MBOAT7 is a lysophosphatidylinositol acyltransferase with remarkable specificity for arachidonoyl-CoA. MBOAT5 and MBOAT7 are particularly susceptible to inhibition by thimerosal. Human neutrophils express mRNA for these four enzymes, and neutrophil microsomes incorporate arachidonoyl chains into phosphatidylinositol, phosphatidylcholine, PS, and phosphatidylethanolamine in a thimerosal-sensitive manner. These results strongly implicate MBOAT5 and MBOAT7 in arachidonate recycling, thus regulating free arachidonic acid levels and leukotriene synthesis in neutrophils.
The human neutrophil is a critically important cell involved in host defense reactions. Part of the inflammatory response mounted by the neutrophil involves generation of leukotriene B4 (LTB4),5 a metabolite of arachidonic acid (20:4) generated through the 5-lipoxygenase pathway. Synthesis of leukotrienes in the neutrophil is a highly complex process under considerable regulation. The initiating event involves elevation of free calcium ion concentration in the cytosol, translocation of cytosolic phospholipase A2 α (cPLA2α) and 5-lipoxygenase to perinuclear membranes, liberation of 20:4 mediated by cPLA2α, association of 5-lipoxygenase with its activating protein, and initiation of 20:4 oxygenation (1). We have reported that inhibition of 20:4 reacylation with thimerosal leads to a greater than 50-fold increase in LTB4 production in human neutrophils, revealing the importance of 20:4 reacylation in limiting the availability of free 20:4 as a part of the regulation of eicosanoid biosynthesis within this cell type (2). The nature and identity of the lysophospholipid acyltransferase (or acyltransferases) inhibited by thimerosal has not been elucidated, yet it has been known for a number of years that the reacylation of free 20:4 into phospholipids by the Lands pathway is quite active in the human neutrophil (3, 4).
Recent investigations have identified several new lysophospholipid acyltransferases (5–10), including some members of the membrane-bound O-acyltransferase (MBOAT) family such as human mboa-7 (henceforth referred to as MBOAT7 in this work), human MBOAT5, and mouse MBOATs 1, 2, and 5. Human MBOAT5 and MBOAT7 have been shown to use 20:4-CoA as a substrate. However, there have not been any reports as to whether or not they are inhibited by low concentrations of thimerosal, a characteristic of the human neutrophil lysophospholipid acyltransferase involved in LTB4 production (2). In related work, our group as well as others (11–15) have recently described a major lysophospholipid acyltransferase in yeast that we refer to as Ale1p. Yeast strains lacking this gene (ale1Δ) are essentially devoid of lysophosphatidylcholine (lyso-PC), lysophosphatidylethanolamine (lyso-PE), lysophosphatidylserine (lyso-PS), and lysophosphatidylglycerol (lyso-PG) acyltransferase activities and have drastically reduced lysophosphatidic acid (lyso-PA) and lysophosphatidylinositol (lyso-PI) acyltransferase activities. Thus, the ale1Δ strain is an ideal host for expression and assay of candidate acyltransferase genes from human and other organisms.
In the present study, mRNAs for MBOATs 1, 2, 5, and 7 were detected in neutrophils, and acyltransferase activities in neutrophil microsomes were found to be consistent with the activities of several MBOAT enzymes. These four members of the human MBOAT family were also expressed in yeast and assayed with a novel LC/MS/MS-based approach, producing an extensive profile of the substrate specificities and thimerosal sensitivity for each enzyme. These results provide strong evidence that MBOATs play a critical role in the regulated synthesis of LTB4 in the human neutrophil.
EXPERIMENTAL PROCEDURES
Materials—Unless otherwise noted, all chemicals, solvents, and amino acids for media were purchased from Sigma or Fisher. Yeast extract, peptone, and yeast nitrogen base were from Difco. Silica-60 TLC plates were from EM Sciences. All lipids were purchased from or donated by Avanti Polar Lipids (Alabaster, AL) except acyl-CoA substrates, which were from Sigma. Synthesis of radiolabeled lyso-PC was performed as described previously (14). PCR primers were from Integrated DNA Technologies (Coralville, IA). The pYES2.1-TOPO kit and anti-V5 antibody were from Invitrogen.
Identification of Human Lysophospholipid Acyltransferases—To find functional orthologs of the yeast Ale1p acyltransferase encoded by the human genome, we used BLASTP to search the human RefSeq protein data base. Four genes with E values of less than 10-10 were members of the poorly characterized family of membrane-bound O-acyltransferase domain-containing proteins: MBOAT1, MBOAT2, MBOAT5, and MBOAT7. The presence of mRNA for these genes in total RNA isolated from human neutrophils was detected by reverse transcription-PCR. Two independent preparations of cDNA from human neutrophils were kindly provided by Dr. Kenneth C. Malcolm (National Jewish Health). Primer pairs used were: MBOAT1: Fwd, 5′-TGAAGTTGCTGGAGGTGAACTGGA-3′; and Rev, 5′-GCCTTTGCGTATGAGCTTGTGGTT-3′; MBOAT2: Fwd, 5′-ATGGGCTGCTTGGCTTCTCATTTG-3′; and Rev, 5′-ACCGCAGTATTTGGAGATGGCTCT-3′; MBOAT5: Fwd, 5′-ACTGGCACCATTGCCTCATTCAAC-3′; and Rev, 5′-ACAGGCCACCAGGGAAATGGATTA-3′; and MBOAT7: Fwd, 5′-ACAGCTACTGCTACGTGGGAATCA-3′; and Rev, 5′-TGTAGTCATAGGCGCGCATCTTCA-3′. The correct size of the products was verified by agarose gel electrophoresis and staining with ethidium bromide.
Cloning and Expression of Human MBOAT Proteins in the Yeast ale1Δ Mutant—Plasmids containing cDNA coding for human MBOAT1 (GenBank™ accession number NM_ 001080480.1), MBOAT2 (GenBank accession number NM_138799.2), and MBOAT5 (GenBank accession number NM_005768) were purchased from Origene (Rockville, MD), and a cDNA clone for MBOAT7 (GenBank accession number AW732353) was purchased from Open Biosystems (Huntsville, AL). The open reading frames were subcloned into the pYES2.1-TOPO vector according to the manufacturer's instructions. Primers used in amplification of the open reading frames were as follows: MBOAT1: Fwd, 5′-ATGGCAGCAGAGCCGCA-3′; and Rev, 5′-ATCTGTTTTTCTCTTATTAATAGAGTTCAG-3′; MBOAT2: Fwd, 5′-ATGGCCACCACCAGCAC-3′; and Rev, 5′-CTGCTTTAGTGATGAATGTCTCGAG-3′; MBOAT5: Fwd, 5′-ATGGCGTCCTCAGCGG-3′; and Rev, 5′-GTCCATCTTCTTTAACTTCTCTTTCCTT-3′; and MBOAT7: Fwd, 5′-ATGTCGCCTGAAGAATGGA-3′; and Rev, 5′-CTCCTCCCGGAGCTTCTCC-3′. The native start codon is underlined in the forward primer, and the stop codon was omitted in the reverse primer, giving an in-frame C-terminal fusion to the V5-His6 epitope tag encoded by the pYES2.1-TOPO vector. Clones bearing inserts with the correct orientation were identified by diagnostic restriction digestions, and several independent plasmids with correct orientation of the insert were transformed into the yeast strain BY4742 ale1Δ::KanMX (16) by standard procedures (17). 2-ml cultures of these transformants were grown to midlog phase (A550 = 0.4–0.8) in synthetic complete glucose medium lacking uracil followed by harvesting of the cells and transfer to 5 ml of synthetic complete galactose medium without uracil for 16 h to induce protein expression. Homogenates of these cultures were prepared by cell disruption with glass beads as described previously (18), and 30 μg of total homogenate protein was subjected to SDS-PAGE and immunoblotting using the mouse monoclonal anti-V5 antibody (at a 1:5000 dilution) to detect the epitope tag. Immunoblot bands were quantified by scanning in film mode on an Epson Perfection 4990 PHOTO scanner. Image analysis and densitometry were performed with the open source National Institutes of Health ImageJ software package (19). Known quantities of a V5-His6-tagged lipid-binding protein from yeast were used for standardizing signal intensities. For biochemical studies, at least two independently isolated clones showing a strong protein expression signal of the correct molecular weight were selected for each gene. Correct gene cloning was verified by DNA sequencing, and yeast strains harboring these plasmids were used for further experiments. To produce membranes for biochemical characterization, expression culture volumes of 100–500 ml were used. Microsomes were prepared as described previously (15) and used for radiochemical and LC/MS/MS-based enzymatic characterization as described below. In all experiments, membranes from yeast transformed with the isogenic empty vector were used as a control.
Acyl-CoA Choice Lyso-PC Acyltransferase Assay—Lyso-PC: acyl-CoA acyltransferase activity was determined by a modification of a previously published LC/MS/MS-based assay (2, 14). Briefly microsomes from yeast (20 μg of protein) were incubated for 30 min at 37 °C in 200 μl of medium (10 mm Tris/HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA) containing 12.5 μm bovine serum albumin, 50 μm [2H31]16:0-lyso-PC, and 30 μm each of eight different acyl-CoA esters with the following acyl groups: 10:0, 14:0, 16:0, 16:1, 18:0, 18:1, 18:2, and 20:4. Reactions were stopped with 750 μl of methanol/chloroform (2:1, v/v), and products were extracted by the Bligh and Dyer (20) method, dried under N2, resuspended in 60 μl of high performance liquid chromatography solvent A (1 mm ammonium acetate in methanol/acetonitrile/water 60:20:20, v/v/v), and injected into the LC/MS/MS system. A gradient from 100% solvent A to 100% solvent B (1 mm ammonium acetate in methanol) was used to elute a C18 column (Columbus 150 × 2 mm, 5 μm, Phenomenex) at a flow rate of 200 μl/min. Solvent B was increased from 0 to 100% in 42 min and kept at 100% for 23 min before re-equilibration. Identification of the phospholipids was carried out using a Sciex API 2000 triple quadrupole mass spectrometer in the positive ion mode with multiple reaction monitoring of the following specific transitions: m/z 681 → 184 for [2H31]16:0/10:0-PC, m/z 737 → 184 for [2H31]16:0/14:0-PC, m/z 765 → 184 for [2H31]16:0/16:0-PC, m/z 763 → 184 for [2H31]16:0/16:1-PC, m/z 793 → 184 for [2H31]16:0/18:0-PC, m/z 791 → 184 for [2H31]16:0/18:1-PC, m/z 789 → 184 for [2H31]16:0/18:2-PC, m/z 813 → 184 for [2H31]16:0/20:4-PC, and m/z 758 → 184 for endogenous 34:2-PC. The amounts of the different phosphatidylcholines were calculated as the percentage of the integrated areas of the corresponding intensity peaks relative to the area of endogenous 34:2-PC.
Dual Substrate (Acyl-CoA and Lysophospholipid) Choice Acyltransferase Assay—Acyltransferase activity was determined essentially as described above using microsomes from yeast or microsomes from human neutrophils obtained as described previously (2). The reaction mixture included the eight acyl-CoAs (30 μm each) as well as 10 μm each of 16:0-lyso-PA, [2H31]16:0-lyso-PC, 16:0-lyso-PE, 18:1-lyso-PG, 17:1-lyso-PI, and 18:1-lyso-PS. After stopping the reaction with 750 μl of methanol/chloroform (2:1, v/v), the following internal standards (5–25 ng each) were added: 17:0/14:1-PA, 17:0/20:4-PC, 17:0/20:4-PE, 17:0/20:4-PG, 17:0/20:4-PI, and 17:0/20:4-PS. Phospholipids were extracted by the Bligh and Dyer (20) method. Samples were resuspended in 75% high performance liquid chromatography solvent C (hexanes/isopropanol 30:40, v/v), 25% solvent D (1 mm ammonium acetate in hexanes/isopropanol/water 30:40:7, v/v/v) and injected into the LC/MS/MS system. A solvent gradient was used to elute a silica column (Prodigy 150 × 1 mm, 5 μm, Phenomenex) at a flow rate of 50 μl/min. Solvent D was maintained at 25% for 5 min, gradually increased to 60% in 10 min and then to 95% in 5 min, and held for 20 min prior to re-equilibration. Identification of the phospholipids was carried out using a Sciex API 3000 triple quadrupole mass spectrometer in the negative ion mode with multiple reaction monitoring of the m/z transitions shown in Table 1. The amounts of the different phospholipid molecular species were measured by calculating the ratio of the integrated area of the corresponding intensity peaks of the analytes to the integrated areas of the peaks corresponding to the internal standard for the same phospholipid class. Area integration and calculations were performed using a Beta version (1.1) of MultiQuant software from Applied Biosystems/MDS Analytical Technologies.
TABLE 1.
Multiple reaction monitoring settings used to detect phospholipids in the dual substrate choice acyltransferase assay Collision-induced m/z transitions, from the molecular ion [M – H]– to the carboxylate ion corresponding to the sn-1 position, monitored for the different phospholipid substrates (lysophospholipids), internal standards, and potential products are shown.
| Class and species | m/z |
|---|---|
| PA | |
| 16:0/OHa | 409 → 255 |
| 17:0/14:1b | 631 → 269 |
| 16:0/10:0 | 563 → 255 |
| 16:0/14:0 | 619 → 255 |
| 16:0/16:0 | 647 → 255 |
| 16:0/16:1 | 645 → 255 |
| 16:0/18:0 | 675 → 255 |
| 16:0/18:1 | 673 → 255 |
| 16:0/18:2 | 671 → 255 |
| 16:0/20:4 | 695 → 255 |
| PC | |
| [2H31]16:0/OHa | 585c → 286 |
| 17:0/20:4b | 854c → 269 |
| [2H31]16:0/10:0 | 739c → 286 |
| [2H31]16:0/14:0 | 795c → 286 |
| [2H31]16:0/16:0 | 823c → 286 |
| [2H31]16:0/16:1 | 821c → 286 |
| [2H31]16:0/18:0 | 851c → 286 |
| [2H31]16:0/18:1 | 849c → 286 |
| [2H31]16:0/18:2 | 847c → 286 |
| [2H31]16:0/20:4 | 871c → 286 |
| PE | |
| 16:0/OHa | 452 → 255 |
| 17:0/20:4b | 752 → 269 |
| 16:0/10:0 | 606 → 255 |
| 16:0/14:0 | 662 → 255 |
| 16:0/16:0 | 690 → 255 |
| 16:0/16:1 | 688 → 255 |
| 16:0/18:0 | 718 → 255 |
| 16:0/18:1 | 716 → 255 |
| 16:0/18:2 | 714 → 255 |
| 16:0/20:4 | 738 → 255 |
| PG | |
| 18:1/OHa | 509 → 281 |
| 17:0/20:4b | 783 → 269 |
| 18:1/10:0 | 663 → 281 |
| 18:1/14:0 | 719 → 281 |
| 18:1/16:0 | 747 → 281 |
| 18:1/16:1 | 745 → 281 |
| 18:1/18:0 | 775 → 281 |
| 18:1/18:1 | 773 → 281 |
| 18:1/18:2 | 771 → 281 |
| 18:1/20:4 | 795 → 281 |
| PI | |
| 17:1/OHa | 583 → 267 |
| 17:0/20:4b | 871 → 269 |
| 17:1/10:0 | 737 → 267 |
| 17:1/14:0 | 793 → 267 |
| 17:1/16:0 | 821 → 267 |
| 17:1/16:1 | 819 → 267 |
| 17:1/18:0 | 849 → 267 |
| 17:1/18:1 | 847 → 267 |
| 17:1/18:2 | 845 → 267 |
| 17:1/20:4 | 869 → 267 |
| PS | |
| 18:1/OHa | 522 → 281 |
| 17:0/20:4b | 796 → 269 |
| 18:1/10:0 | 676 → 281 |
| 18:1/14:0 | 732 → 281 |
| 18:1/16:0 | 760 → 281 |
| 18:1/16:1 | 758 → 281 |
| 18:1/18:0 | 788 → 281 |
| 18:1/18:1 | 786 → 281 |
| 18:1/18:2 | 784 → 281 |
| 18:1/20:4 | 808 → 281 |
Lysophospholipid substrate
Internal standard
The ions monitored for PC are acetate adducts, [M + OAc]–
Radiochemical Assays of MBOAT5—Lyso-PC, lyso-PE, and lyso-PS acyltransferase activities of MBOAT5 were measured essentially as described previously for the yeast Ale1p enzyme (14). Standard acyltransferase assays were conducted in 200-μl final volume, and buffer and substrate compositions were 50 μm 18:1-lysophospholipid, 50 μm acyl-CoA, 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, and 10,000–20,000 cpm [14C]18:1-lysophospholipid. Reactions were initiated by addition of 30 μg of yeast membrane protein, and incubation proceeded for 0.5–1 h at 30 °C. Reactions were terminated by Bligh and Dyer (20) extraction, and the products were resolved by thin-layer chromatography on Silica-60 plates in the solvent chloroform/methanol/water (65:25:5, v/v/v) followed by quantification of mono- and diacyl lipids using a PhosphorImager. Membranes prepared from the ale1Δ pYES2.1 vector-transformed control strain were included as a background control, and this background was subtracted from the counts present in MBOAT5-containing assays. The residual activity in the control membranes was always <5% of the value for the MBOAT5 membranes. In some experiments, MBOAT5-containing membranes were treated with 50 μm thimerosal for 15 min at 30 °C prior to assay.
Statistical Analysis—Where indicated, results were analyzed by an unpaired t test using GraphPad software.
RESULTS
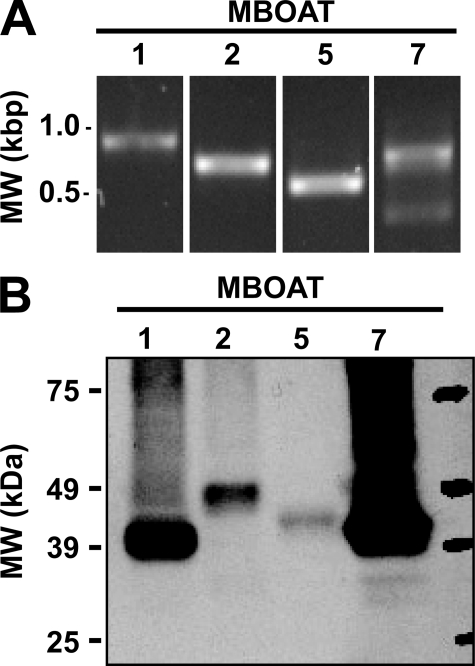
Expression of mRNA for Putative Human Orthologs of Yeast Ale1p in Human Neutrophils—Our laboratory (14, 15) and several other groups (11–13) have recently reported that Ale1p is the major lysophospholipid acyltransferase in Saccharomyces cerevisiae. The sequence of yeast Ale1p was used to search the human protein data base for similar sequences. The most closely related proteins were MBOAT1, MBOAT2, and MBOAT5 for which no enzymatic activities had been described at the time these experiments were started. Another closely related gene, MBOAT7 (also known as LRC4, LENG4, and BB1), has recently been identified and characterized as a lyso-PI acyltransferase in an interesting study by Lee et al. (21), who proposed the name mboa-7 for the Caenorhabditis elegans gene. In addition, the enzymatic activities of murine MBOAT1, MBOAT2, and MBOAT5 have recently been reported (6). The expression of MBOAT mRNA in human neutrophils was determined by PCR. As shown in Fig. 1A, PCR products of the appropriate size for MBOAT1, MBOAT2, MBOAT5, and MBOAT7 were amplified from total RNA from human neutrophils. Because any of the aforementioned MBOATs could potentially be involved in 20:4 reacylation in neutrophils, all four constructs were cloned and expressed in yeast to study their enzymatic properties.
FIGURE 1.
Expression of MBOAT mRNAs in human neutrophils and production of human MBOAT proteins in ale1Δ yeast. A, expression of human MBOAT mRNAs was examined using cDNA prepared from total human neutrophil RNA by PCR with primer pairs specific for MBOAT1 (expected 795 bp), MBOAT2 (expected 611 bp), MBOAT5 (expected 495 bp), and MBOAT7 (expected 765 bp). Reaction products and DNA size standards (left lane) were visualized by ethidium bromide staining after agarose gel electrophoresis. B, 30 μg of total microsomal protein from ale1Δ yeast expressing V5-His6-tagged human MBOAT proteins were analyzed by immunoblotting with an anti-V5 antibody. Data shown are representative of at least two independent microsome preparations.
Cloning and Expression of Human MBOAT Acyltransferases in Yeast—The MBOAT open reading frames were cloned into a yeast expression vector with a C-terminal fusion to a V5-His6 epitope/purification tag encoded by the vector. These constructs were then transformed into an ale1Δ strain of yeast. The full-length proteins were well expressed as determined by the presence of a ∼50-kDa signal on immunoblots using an antibody against the V5 epitope (Fig. 1B). All of these MBOAT proteins were predicted to contain six to eight transmembrane helices by the program TMHMM (22, 23), and in agreement with this, the proteins were exclusively present in membrane preparations and were absent from the cytosolic fractions (100,000 × g supernatant). Two independent preparations were analyzed by immunoblotting and densitometry, which yielded an average MBOAT acyltransferase expression (per 30 μg of total microsomal protein) of 47 (MBOAT1), 11 (MBOAT2), 5 (MBOAT5), and 165 ng (MBOAT7). These values were used to normalize acyltransferase activity assays when comparing the different enzymes.
Lyso-PC:Acyl-CoA Acyltransferase Activities of MBOAT Enzymes—In an approach similar to that used to characterize the lyso-PC acyltransferase activity of Ale1p, microsomes from yeast expressing the human MBOAT proteins were initially tested for lyso-PC acyltransferase activity using 1-[2H31]palmitoyl-2-hydroxyl-sn-glycero-3-phosphocholine (deuterated lyso-PC) and a mixture of eight fatty acyl-CoAs to study the substrate specificity of these enzymes when a choice of substrates is available (14). Products were analyzed by LC/MS/MS. As shown in Fig. 2, there was very little activity observed with membranes from ale1Δ yeast bearing the empty vector; the main product of this residual activity was [2H31]16: 0/16:1-PC. This finding shows that yeast express an enzyme, other than Ale1p, that is able to incorporate palmitoleoyl-CoA into PC albeit much less efficiently. The result also highlights the usefulness of the ale1Δ yeast as a low background model to study heterologous acyltransferases. The most abundant PC detected in these samples was non-deuterated 34:2-PC, most likely 16:1/18:1-PC present in yeast microsomes. The LC/MS/MS signal of this endogenous lipid showed some small variation between different microsomal preparations, but it provided a useful internal standard to normalize the signals of the reaction products.
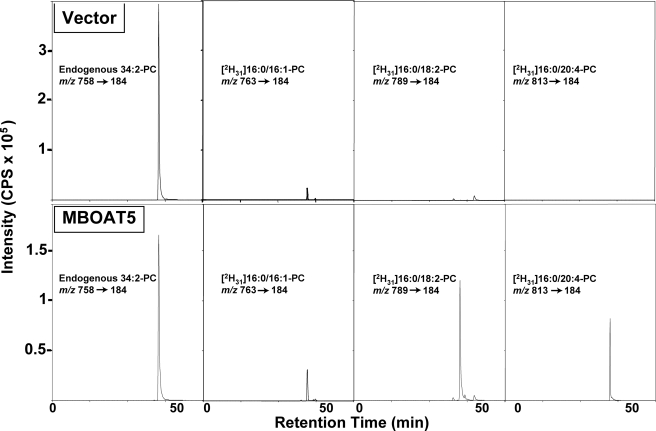
FIGURE 2.
Human MBOAT5 is a human lyso-PC acyltransferase with preference for linoleoyl- and arachidonoyl-CoA. A lyso-PC acyltransferase assay was performed with microsomes from yeast transformed with either an empty vector (top panels) or vector containing the open reading frame for human MBOAT5 (bottom panels) in a reaction containing deuterated 16:0-lyso-PC and a mixture of acyl-CoAs as substrates. The different species of deuterated PC formed during the assay were analyzed by lipid extraction and LC/MS/MS monitoring of specific collision-induced m/z transitions. Panels show the complete chromatographic profile of the ion intensities of the transitions for endogenous 34:2-PC or the deuterated PCs resulting from the incorporation of palmitoleoyl (16:1), linoleoyl (18:2), or arachidonoyl (20:4) acyl chains. Results are from one experiment representative of three experiments performed with independent microsome preparations. cps, counts/s.
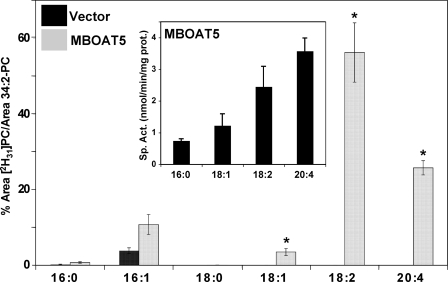
Microsomes from yeast expressing human MBOAT5 catalyzed increased production of 16:1- and 18:1-containing PC but especially pronounced production of PC containing 18:2 and 20:4 acyl chains. MBOAT5 exhibited very little activity with saturated acyl-CoAs containing 16:0 and 18:0 acyl chains (and also 10:0 and 14:0; not shown). The Fig. 3, inset, shows that the results of the choice assay are in good agreement with parallel assays of MBOAT5-containing microsomes with radioactive lysophospholipid and a single acyl-CoA. With either type of assay, the highest activity was with linoleoyl- and arachidonoyl-CoA. Under optimal conditions with a single acyl-CoA substrate, the enzyme used 20:4-CoA maximally with a turnover number (kcat) of ∼700 s-1. The substrate specificity of MBOAT5 was very similar between microsomes and whole cell lysates (data not shown), indicating that the presence of cytosolic proteins does not significantly affect the specificity of the enzyme for polyunsaturated acyl-CoAs.
FIGURE 3.
Human MBOAT5 exhibits preference for polyunsaturated acyl-CoAs. Acyltransferase assays were performed as described for Fig. 2. The amount of each individual deuterated PC species is expressed as the percentage of its LC/MS/MS intensity integrated area compared with the endogenous 34:2 PC intensity integrated area. Results shown represent average ± S.E. (vector, n = 3; MBOAT5, n = 6). Inset, acyltransferase activity of MBOAT5-expressing yeast microsomes was measured with radiolabeled 18:1-lysoPC and individual acyl-CoA species containing the indicated fatty acyl chains in separate reactions. The data represent the average ± S.D. of three separate experiments conducted in duplicate. *, p < 0.05 (compared with empty vector).
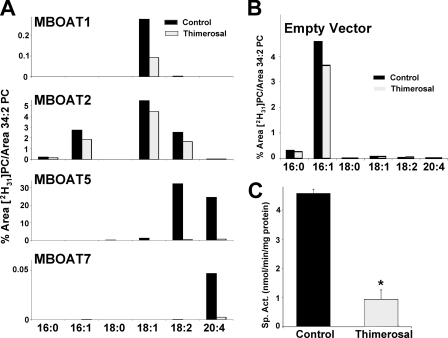
Thimerosal Sensitivity of MBOAT Lyso-PC Acyltransferase Activities—Acyltransferase activities of the four MBOAT proteins were analyzed in the presence or absence of the organomercury compound thimerosal (Fig. 4A). To better appreciate the contribution of each enzyme, the activity of microsomes from control yeast (empty vector; Fig. 4B) was subtracted from each one of the panels in Fig. 4A. Activities were also normalized for MBOAT expression levels as assessed by densitometry (see Fig. 1B). MBOAT1 showed very weak lyso-PC acyltransferase activity using 18:1-CoA preferentially. MBOAT7 also had weak but detectable and reproducible activity with 20:4-CoA. MBOAT2 exhibited higher activity with preference for 16:1-, 18:2-, and especially 18:1-CoA but no measurable activity toward 20:4-CoA. The use of 50 μm thimerosal revealed interesting differences between MBOAT activities in vitro. The residual acyltransferase activities present in control yeast (Fig. 4B), as well as MBOAT1 and MBOAT2, were only marginally inhibited by thimerosal. In contrast, thimerosal profoundly inhibited the acyltransferase activities of MBOAT5 and MBOAT7. The radiochemical assay data (Fig. 4C) confirmed that the MBOAT5-dependent lyso-PC:20:4-CoA acyltransferase activity is strongly inhibited by pretreatment of microsomes with 50 μm thimerosal even in an assay under optimal conditions with only 20:4-CoA available to the enzyme.
FIGURE 4.
Human MBOATs 5 and 7 show arachidonoyl-CoA selectivity and thimerosal sensitivity. An acyl-CoA substrate choice acyltransferase assay was performed with microsomes from yeast transformed with empty vector or plasmids expressing the indicated proteins in the absence or presence of 50 μm thimerosal. The amount of each individual deuterated PC species is expressed as the percentage of its LC/MS/MS signal relative to the endogenous 34:2 PC signal. A, microsomes from yeast expressing human MBOAT proteins. Data are normalized for MBOAT expression, and background activity present in yeast transformed with empty vector is subtracted. This background activity is shown in B. Data shown are from one experiment representative of two individual experiments. C, MBOAT5 acyltransferase assays conducted using radiolabeled lyso-PC and arachidonoyl-CoA either in the absence or in the presence of 50 μm thimerosal. Data shown are the average ± S.E. of two individual experiments conducted in duplicate. *, p < 0.005 (compared with control).
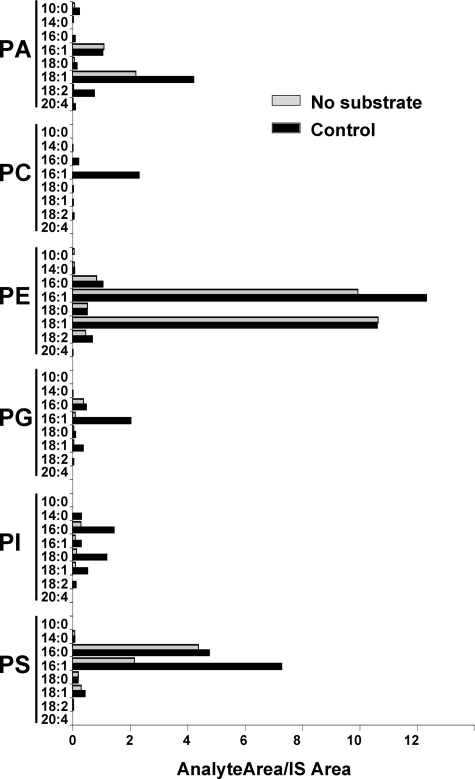
Lysophospholipid Substrate Specificity and Thimerosal Sensitivity of MBOAT Acyltransferases—The activities of murine acyltransferases MBOAT1, MBOAT2, and MBOAT5 have been recently studied in detail, and data are available regarding their substrate specificities using radioactive lysophospholipids (6). However, no comprehensive data are available about the human homologs. We used a novel, LC/MS/MS-based assay to directly determine the selectivity of acyltransferases in reactions containing a mixture of different lysophospholipids, as well as the different acyl-CoAs used in lyso-PC acyltransferase reactions, as described above. These assays were also carried out in the presence or absence of 50 μm thimerosal. Microsomes from Ale1p-deficient yeast transformed with empty vector showed weak acyltransferase activity for all classes of phospholipids (Fig. 5), adding support to the notion that yeast contain minor acyltransferases in addition to the major acyltransferase Ale1p. Reactions containing microsomes but no acyl-CoAs or lysophospholipids were analyzed as a phospholipid background control. Under these conditions, we detected the presence of endogenous phospholipids, such as 16:1- and 18:1-containing PA and PE or 16:0- and 16:1-containing PS among others. It is important to note that this is not a comprehensive profile of the phospholipids present in yeast microsomes but rather an evaluation of the possible phospholipid background that could interfere with the interpretation of the enzymatic activity data. This control proved unnecessary when using deuterated or odd chain lysophospholipid substrates as shown by the negligible signal present in the PC and PI classes, respectively. For the other lipid classes, acyltransferase activities were clearly detectable above background for PA (mainly 18:1 and 18:2), PE (16:1), PG (16:1), PI (16:0, 18:0, and 18:1), and PS (16:1). Interestingly the lyso-PC activity with 16:1 CoA, which is shown in Fig. 4B, was also detectable in this dual substrate choice assay. None of these acyltransferase activities present in yeast were substantially inhibited by 50 μm thimerosal (data not shown).
FIGURE 5.
Dual substrate choice assay of lysophospholipid acyltransferases present in ale1Δ yeast microsomes. Microsomes from yeast transformed with empty vector were incubated with a mixture of eight acyl-CoAs (30 μm each) as detailed above and a mixture of six lysophospholipids (lyso-PA, deuterated lyso-PC, lyso-PE, lyso-PG, lyso-PI, and lyso-PS; 10 μm each). The amount of each individual phospholipid species formed in the assay is expressed as the ratio of its integrated LC/MS/MS signal relative to the corresponding phospholipid internal standard (IS) added at the end of the incubation. Data shown are from one experiment representative of two independent experiments.
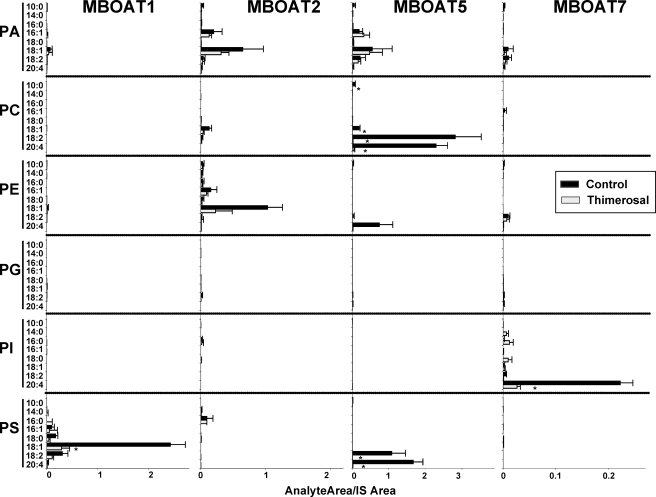
In Fig. 6, the activity of all four MBOATs studied in the dual substrate choice assay (containing six lysophospholipids and eight acyl-CoAs) is shown. The background activity present in control yeast microsomes (Fig. 5) was subtracted from the data obtained with microsomes from yeast expressing human MBOAT enzymes, so the activity contributed by each individual enzyme is easier to appreciate. In the case of MBOAT1, lyso-PS was the major acyl acceptor, mainly with 18:1-CoA and to a lesser extent 18:2-CoA. This activity was inhibited by thimerosal by about 75%. MBOAT2 was mostly active incorporating 18:1 acyl chains into PE and PA. The activity of MBOAT2 with lyso-PC (mainly incorporating 18:1-CoA) described in Fig. 4A was also detected, however, at a level much lower than that with lyso-PE and lyso-PA. MBOAT2 activity was also partially inhibited by thimerosal. As expected, the main activity of MBOAT5 used lyso-PC as acyl acceptor, and consistent with the results shown above, 18:2-CoA and 20:4-CoA were used preferentially as acyl donors. Interestingly MBOAT5 also incorporated these same acyl-CoAs into PS and to a lesser extent into PE. MBOAT5 activity was completely abrogated by thimerosal. In the case of MBOAT7, the major activity observed was with lyso-PI and with 20:4-CoA as substrates. Thimerosal almost completely inhibited this activity. In this case, no activity of MBOAT7 incorporating 20:4 into PC was detected, indicating that, relative to lyso-PI, lyso-PC is not a good acyl acceptor for the activity of MBOAT7 and that this minor lyso-PC acyltransferase activity then becomes undetectable.
FIGURE 6.
Dual substrate choice acyltransferase assays demonstrate the preferential utilization of arachidonic acid and thimerosal sensitivity of MBOATs 5 and 7. Acyltransferases were assayed as described above with microsomes from yeast expressing the indicated human proteins either in the absence or in the presence of 50 μm thimerosal. The amount of each individual phospholipid species is expressed as the ratio of its integrated LC/MS/MS signal relative to the corresponding phospholipid internal standard (IS) added at the end of the incubation. Data are normalized for MBOAT expression, and background activity present in yeast transformed with empty vector is subtracted. Data shown are average ± S.E. of two individual experiments performed in triplicate. *, p < 0.05 (compared with control).
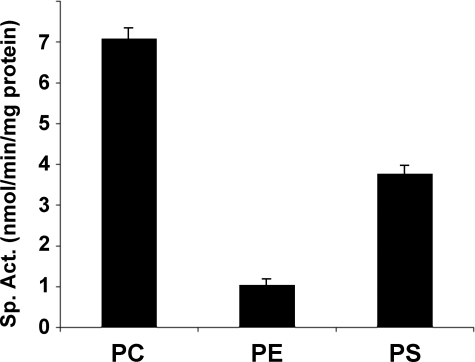
The MBOAT5 substrate specificity observed in this dual choice assay has potentially profound implications for arachidonate recycling. Because of this, radiochemical assays using 20:4-CoA and lyso-PC, lyso-PS, or lyso-PE individually were performed. Under these conditions, MBOAT5 exhibited specificity and catalytic activity very similar to that found in the dual choice assay. The results shown in Fig. 7 confirmed that MBOAT5 exhibits preferences for 20:4-CoA and lysophospholipids with a rank order of PC > PS > PE, which is identical to that seen with the dual choice assay.
FIGURE 7.
Human MBOAT5 acylates the lysoaminoglycerophospholipids. Acyltransferase assays were conducted with arachidonoyl-CoA using radiolabeled lyso-PC, lyso-PE, or lyso-PS (all substrates at 50 μm). Data shown represent the average ± S.E. of three individual experiments conducted in duplicate.
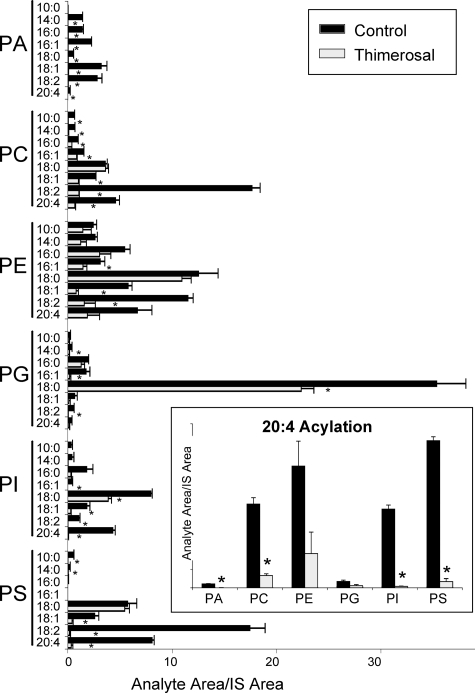
Lysophospholipid Acyltransferase Activities in Microsomes from Human Neutrophils—Our interest in lysophospholipid acyltransferases was stimulated because of the effect of arachidonic acid reacylation on eicosanoid synthesis in human neutrophils (2). We used our newly developed dual substrate choice assay to study the reacylation activities of neutrophil microsomes when presented with a mixture of lysophospholipids and acyl-CoAs. As shown in Fig. 8, the observed pattern of activities is consistent with the action of most, if not all, MBOAT acyltransferases. Reacylation activities were detected in the six classes of phospholipids studied with diverse acyl chain specificities. The background signals from endogenous phospholipids detected in microsomes incubated in the absence of exogenous lysophospholipids or acyl-CoAs were subtracted from the data shown. The most prominent activity was the incorporation of stearoyl chains into PG. This is not an activity displayed by any of the MBOATs studied here and may be due to lysophosphatidylglycerol acyltransferase 1 (LPGAT1), a human enzyme that shows strong preference for lyso-PG and 18:0-CoA and is widely expressed, including in cells in peripheral blood (24). Other major activities observed incorporated linoleate into PC, PE, and PS and could be accounted for by the action of MBOAT5. Incorporation of stearate into PE, PI, and PS was also prominent. Not surprisingly, one conclusion from this experiment is that additional acyltransferases exhibiting high specific activity and with substrate specificities distinct from the MBOATs exist in the human neutrophil. However, expression of MBOAT5 and MBOAT7 would be sufficient to explain the 20:4-CoA acyltransferase activities (Fig. 8, inset). Interestingly 20:4 chains were introduced at similar rates into PC, PE, PI, and PS with small but detectable activities observed for PA and PG. These data demonstrate that, most likely, there is not one single acyltransferase that is responsible for 20:4 recycling into phospholipids in human neutrophils but rather that different enzymes contribute to this essential step in maintaining low levels of free arachidonic acid in these cells. More specifically, these data identify a major role for MBOAT5 (most likely responsible for 20:4 incorporation into PC, PE, and PS) and for MBOAT7 (most likely responsible for incorporation into PI) in regulating the amount of free 20:4 available for ecosanoid production. This idea is also supported by the fact that thimerosal greatly inhibits incorporation of 20:4 into phospholipids in the human neutrophil and that both MBOAT5 and MBOAT7 are markedly inhibited by this compound.
FIGURE 8.
Human MBOATs 5 and 7 are the dominant arachidonoyl-CoA-selective and thimerosal-sensitive acyltransferases of human neutrophils. Dual substrate choice acyltransferase assays of microsomes from human neutrophils were performed as described above either in the absence or in the presence of 50 μm thimerosal. The amount of each individual phospholipid species is expressed as the ratio of its integrated LC/MS/MS signal relative to the corresponding phospholipid internal standard (IS) added at the end of the incubation. For clarity, the data for arachidonoyl (20:4) incorporation into phospholipids are shown separately in the inset. Data shown are average ± S.E. of two individual experiments performed in triplicate. *, p < 0.05 (compared with control).
DISCUSSION
The biochemical reactions that constitute the phospholipid remodeling cycle were described almost half a century ago by Lands (25). The identities and biochemical characteristics of the PLA2 enzymes involved in the deacylation arm of the cycle have been extensively studied especially as they relate to the release of 20:4 for eicosanoid production (26). The reacylation arm of the cycle, catalyzed by acyl-CoA ligases and lysophospholipid acyltransferases, has also been well studied (3), but the genes and proteins involved in this process have remained largely unknown. However, a renewed interest in this important aspect of phospholipid metabolism has resulted in the recent identification and characterization of a number of genes that encode lysophospholipid acyltransferases. The acyltransferases segregate into two distinct phylogenetic clades. Several researchers have identified PlsC domain-containing proteins, referred to as LPCATs, that are involved in the acylation of lyso-PC in the production of lung surfactant (5, 7), the production of platelet-activating factor (8), and remodeling of the erythrocyte membrane (9). This group also includes a lyso-PE acyltransferase, termed LPEAT2, which is highly expressed in brain and also has some activity with lyso-PG, lyso-PS, and lyso-PC (27). The second group includes the human homologs of the yeast enzyme Ale1p, which was initially identified as a lyso-PE acyltransferase but which is also a major acyltransferase using lyso-PA, lyso-PC, lyso-PG, lyso-PI, and lyso-PS as substrates (14, 15). Mammalian homologs of Ale1p belong to the family of MBOATs. MBOAT5 (also named LPCAT3) has been identified as a lyso-PC acyltransferase that can also use lyso-PE and lyso-PS as substrates (6, 10). Mouse MBOAT1 (also termed LPEAT1) uses lyso-PE and lyso-PS as substrates, whereas mouse MBOAT2 (LPCAT4) exhibits lyso-PC and lyso-PE acyltransferase activities (6). Finally human MBOAT7 (also known as LRC4, LENG4, BB1, or mboa-7) has been shown to exhibit lyso-PI acyltransferase activity (21).
The MBOAT superfamily of membrane-associated proteins includes enzymes such as acyl-CoA:cholesterol acyltransferases 1 and 2, ACAT1 and ACAT2 (28), and diacylglycerol O-acyltransferase, DGA1 (29), as well as the glycosylphosphatidylinositol anchor-remodeling enzyme, Gup1p (30). This family of enzymes is evolutionarily unrelated to the PlsC domain-containing LPCAT1/LPCAT2/LPEAT2 family; therefore we propose that the LPCAT designation be used for this latter family of enzymes because it includes the first identified human lyso-PC acyltransferases while retaining the MBOAT designation for the homologs of yeast Ale1p. This proposed nomenclature may also avoid some confusion given the fact that all of the MBOAT enzymes characterized exhibit some promiscuity regarding the lysophospholipid polar head. It has been suggested that a His residue included in a hydrophobic region (His-374 in MBOAT5) may be a part of the active site as mutation of this residue in Ale1p and MBOAT7 abolishes enzymatic activity (21, 31). All members of this family are predicted to contain several membrane-spanning regions and are therefore thought to be integral membrane proteins. This is consistent with the fact that we observed similar acyltransferase activity and specificity in whole cell lysates and microsome preparations from yeast expressing human MBOAT5 and is also consistent with immunolocalization data (6, 10). Interestingly MBOAT5 has also been recently identified in a high throughput protein function screen as playing a relevant role in Golgi vesicle trafficking in RAW264.7 cells (32). In that study, no enzymatic activity for the protein was described, but an antibody was raised that distinctly stained the endoplasmic reticulum in HeLa and HEK293 cells. These observations raise the possibility of phospholipid remodeling through MBOAT5 playing a role in Golgi apparatus integrity as well as in endoplasmic reticulum-Golgi vesicular traffic.
In the present study, we investigated the role of MBOATs in the thimerosal-sensitive reacylation of 20:4 in neutrophil microsomes. This 20:4 reacylation plays a major role in regulating leukotriene biosynthesis in neutrophils (2). This assertion was supported by the finding that thimerosal profoundly inhibited the lyso-PC:20:4-CoA acyltransferase activity of neutrophil membranes, and treatment of neutrophils with the compound synergistically enhanced leukotriene production induced by granulocyte/macrophage colony-stimulating factor and formylmethionylleucylphenylalanine. The presence of mRNAs for MBOAT proteins in human neutrophils offered the possibility that these enzymes may be involved in 20:4 reacylation and prompted us to investigate their properties.
The recent identification of the yeast Ale1p acyltransferase has provided important new tools for the characterization of putative human acyltransferases from both the LPCAT and MBOAT families. One of these tools, the lysophospholipid acyltransferase-deficient ale1Δ yeast strain, was exploited in this study as a host for the expression of enzymes belonging to the MBOAT family. This heterologous expression strategy has recently been used for the characterization of two plant homologs of Ale1p, AtLPLAT1 and AtLPLAT2 from Arabidopsis thaliana (33). The biochemical data presented in this work provide an extensive evaluation of the lysophospholipid acyltransferase activities of MBOAT enzymes that are expressed in the human neutrophil. This analysis was aided by the use of an acyl-CoA choice assay as published previously (14).
To provide additional information regarding lysophospholipid preferences of these enzymes, we developed a novel dual substrate choice assay that allows for the simultaneous determination of the lysophospholipid and acyl-CoA specificities. This is useful because it allows the parallel analysis of both lysophospholipid and acyl-CoA preferences by simultaneous measurement of a large number (48 in our case) of products from a single reaction. More importantly, it may more closely approximate the cellular environment of enzymes that are exposed to different substrates and that are in a dynamic equilibrium resulting from normal cell metabolism or cellular activation. One caveat of this approach is the biophysical complexity of the experiment. It involves two groups of compounds with detergent properties (lysophospholipids and acyl-CoAs) at combined concentrations that may exceed their individual critical micellar concentrations. The assays also contain microsomal membranes and albumin, both of which have been shown to bind these amphipathic molecules (34), which would raise their apparent critical micellar concentrations. These factors probably affect the relative availability of the individual substrates, and in fact, there are small differences in apparent substrate specificity when the mixed substrate versus single substrate assays are compared. However, our experiments show that this dual substrate choice approach is valid for acyltransferases because the overall qualitative substrate preferences (e.g. 18:2 and 20:4 chains for MBOAT5) are the same for individual substrate experiments (Fig. 3, inset), acyl-CoA choice experiments (Fig. 3), or dual choice experiments (Fig. 6). These results are also consistent with published data about these enzymes with the single possible exception of MBOAT1. Under our experimental conditions, robust lyso-PS acyltransferase activity incorporating oleoyl-CoA was observed, but only very weak lyso-PE acyltransferase activity was observed in contrast with the data of Hishikawa et al. (6), who report both lyso-PE and lyso-PS activities. Although it cannot be ruled out that there are differences in specificity between the human and murine enzymes, this apparent discrepancy could be due to the fact that we used 16:0-lyso-PE and 18:1-lyso-PS. Although the latter has been shown to be a good substrate for MBOAT1, 18:1-lyso-PE is a better substrate than 16:0-lyso-PE, which could reduce lyso-PE acyltransferase activity in our competitive assay conditions. This notion is supported by the results obtained with MBOAT2, which prefers 16:0-lyso-PE to 18:1-lyso-PE (6).
The data with microsomes from human neutrophils show incorporation of arachidonoyl acyl chains into PC, PE, PI, and PS. Because only MBOAT5 (for synthesis of PC, PS, and PE) and MBOAT7 (for synthesis of PI and to a much lesser extent PC) seem to efficiently use 20:4-CoA, these results provide strong evidence that both enzymes are involved in the thimerosal-sensitive 20:4 reacylation in human neutrophils. Among the novel lysophospholipid acyltransferases, LPEAT2 has weak but detectable activity with 20:4-CoA, and LPCAT2, which is a lyso-platelet-activating factor:acetyl-CoA acyltransferase, is also able to use long chain fatty acyl-CoAs, including 20:4-CoA (8). However, preincubation of human neutrophils with thimerosal before granulocyte/macrophage colony-stimulating factor and formylmethionylleucylphenylalanine treatment resulted in an increase in platelet-activating factor production, suggesting that LPCAT2 is either not sensitive to thimerosal or not a limiting step in platelet-activating factor production in neutrophils.6
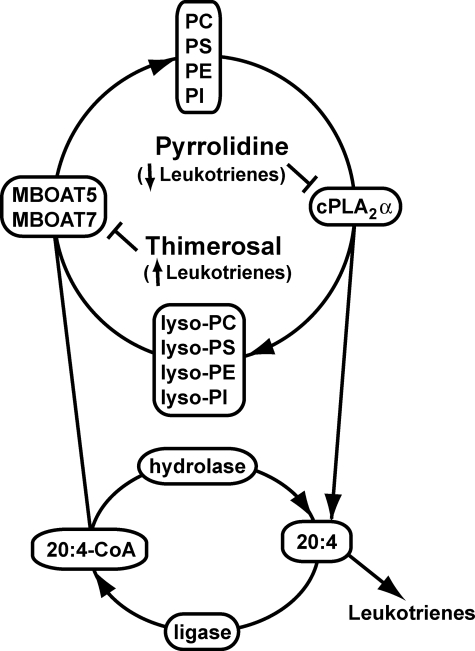
In view of the effect of thimerosal on eicosanoid synthesis and the sensitivity of MBOAT5 and MBOAT7 to this inhibitor, it is intriguing to speculate that the presence of these acyltransferases in intracellular compartments (such as the endoplasmic reticulum) where other proteins involved in eicosanoid synthesis, such as cPLA2α or 5-lipoxygenase, are localized in stimulated cells could play a role in regulating reacylation and availability of free 20:4. However, the actual rate of fatty acid and lysophospholipid reacylation in resting or activated cells is ultimately the result of a variety of factors including expression and activity of enzymes that include phospholipases, acyl-CoA ligases, acyl-CoA hydrolases, CoA-independent transacylases, and acyl-CoA acyltransferases. Therefore, data describing the expression and activity of these enzymes will help greatly in understanding the mechanisms regulating the levels of free fatty acids in cells and therefore eicosanoid production. In Fig. 9, we present a scheme for the role of reacylation and MBOATs in 20:4 recycling and leukotriene production in neutrophils.
FIGURE 9.
Integrated model for phospholipid remodeling and arachidonic acid mobilization in human neutrophils. The major phospholipids in the human neutrophil are PC, PS, PE, and PI. Upon activation of the cell, 20:4 in the sn-2 position of these membrane lipids is released by the action of cPLA2α, generating the corresponding lysophospholipids and free 20:4. 20:4 can then be used for the production of various proinflammatory eicosanoid species (primarily LTB4) or can be returned to the phospholipid pool via the action of an acyl-CoA ligase and either MBOAT7 (that uses lyso-PI) or MBOAT5 (that uses lyso-PC, lyso-PS, and lyso-PE). Pyrrolidine, an inhibitor of cPLA2α, and thimerosal, an MBOAT inhibitor, decrease and increase, respectively, the production of eicosanoids, underscoring the importance of this deacylation/reacylation cycle in regulating the production of lipid-derived inflammatory mediators.
In summary, our data describe the characterization of a group of novel human lysophospholipid acyltransferases. The critical finding that stems from this work is the regulatory role that we have elucidated for MBOAT5 and MBOAT7, which exhibit robust activity using polyunsaturated acyl-CoAs, including arachidonoyl-CoA. These activities, as well as their sensitivity to the ethylmercury compound thimerosal, coupled with the pattern of acyltransferase activities observed in human neutrophils strongly suggest that these enzymes play a major role in regulating arachidonic acid availability for eicosanoid biosynthesis.
Acknowledgments
We are very grateful to Dr. Kenneth Malcolm (National Jewish Health, Denver, CO) for providing human neutrophil cDNA and to Avanti Polar Lipids for the generous gift of phospholipid internal standards.
This work was supported, in whole or in part, by National Institutes of Health Grants GM076798 (to W. R. R.), HL025785 (to R. C. M.), 5R37GM032453 (to D. R. V.), and GM081461 (to D. R. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: LTB4, leukotriene B4; MBOAT, membrane-bound O-acyltransferase; 20:4, arachidonic acid; LC/MS/MS, liquid chromatography coupled to tandem mass spectrometry; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; cPLA2α, cytosolic phospholipase A2 α; Fwd, forward; Rev, reverse; LPCAT, lysophosphatidylcholine acyltransferase.
M. A. Gijón, S. Zarini, and R. C. Murphy, unpublished observation.
References
- 1.Murphy, R. C., and Gijon, M. A. (2007) Biochem. J. 405 379-395 [DOI] [PubMed] [Google Scholar]
- 2.Zarini, S., Gijon, M. A., Folco, G., and Murphy, R. C. (2006) J. Biol. Chem. 281 10134-10142 [DOI] [PubMed] [Google Scholar]
- 3.Chilton, F. H., and Murphy, R. C. (1986) J. Biol. Chem. 261 7771-7777 [PubMed] [Google Scholar]
- 4.Reinhold, S. L., Zimmerman, G. A., Prescott, S. M., and McIntyre, T. M. (1989) J. Biol. Chem. 264 21652-21659 [PubMed] [Google Scholar]
- 5.Chen, X., Hyatt, B. A., Mucenski, M. L., Mason, R. J., and Shannon, J. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11724-11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hishikawa, D., Shindou, H., Kobayashi, S., Nakanishi, H., Taguchi, R., and Shimizu, T. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 2830-2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakanishi, H., Shindou, H., Hishikawa, D., Harayama, T., Ogasawara, R., Suwabe, A., Taguchi, R., and Shimizu, T. (2006) J. Biol. Chem. 281 20140-20147 [DOI] [PubMed] [Google Scholar]
- 8.Shindou, H., Hishikawa, D., Nakanishi, H., Harayama, T., Ishii, S., Taguchi, R., and Shimizu, T. (2007) J. Biol. Chem. 282 6532-6539 [DOI] [PubMed] [Google Scholar]
- 9.Soupene, E., Fyrst, H., and Kuypers, F. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 88-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao, Y., Chen, Y., Bonacci, T. M., Bredt, D. S., Li, S., Bensch, W. R., Moller, D. E., Kowala, M., Konrad, R. J., and Cao, G. (2008) J. Biol. Chem. 283 8258-8265 [DOI] [PubMed] [Google Scholar]
- 11.Benghezal, M., Roubaty, C., Veepuri, V., Knudsen, J., and Conzelmann, A. (2007) J. Biol. Chem. 282 30845-30855 [DOI] [PubMed] [Google Scholar]
- 12.Chen, Q., Kazachkov, M., Zheng, Z., and Zou, J. (2007) FEBS Lett. 581 5511-5516 [DOI] [PubMed] [Google Scholar]
- 13.Jain, S., Stanford, N., Bhagwat, N., Seiler, B., Costanzo, M., Boone, C., and Oelkers, P. (2007) J. Biol. Chem. 282 30562-30569 [DOI] [PubMed] [Google Scholar]
- 14.Riekhof, W. R., Wu, J., Gijon, M. A., Zarini, S., Murphy, R. C., and Voelker, D. R. (2007) J. Biol. Chem. 282 36853-36861 [DOI] [PubMed] [Google Scholar]
- 15.Riekhof, W. R., Wu, J., Jones, J. L., and Voelker, D. R. (2007) J. Biol. Chem. 282 28344-28352 [DOI] [PubMed] [Google Scholar]
- 16.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., Chu, A. M., Connelly, C., Davis, K., Dietrich, F., Dow, S. W., El Bakkoury, M., Foury, F., Friend, S. H., Gentalen, E., Giaever, G., Hegemann, J. H., Jones, T., Laub, M., Liao, H., Liebundguth, N., Lockhart, D. J., Lucau-Danila, A., Lussier, M., M'Rabet, N., Menard, P., Mittmann, M., Pai, C., Rebischung, C., Revuelta, J. L., Riles, L., Roberts, C. J., Ross-MacDonald, P., Scherens, B., Snyder, M., Sookhai-Mahadeo, S., Storms, R. K., Veronneau, S., Voet, M., Volckaert, G., Ward, T. R., Wysocki, R., Yen, G. S., Yu, K., Zimmermann, K., Philippsen, P., Johnston, M., and Davis, R. W. (1999) Science 285 901-906 [DOI] [PubMed] [Google Scholar]
- 17.Gietz, R. D., and Woods, R. A. (2002) Methods Enzymol. 350 87-96 [DOI] [PubMed] [Google Scholar]
- 18.Riekhof, W. R., and Voelker, D. R. (2006) J. Biol. Chem. 281 36588-36596 [DOI] [PubMed] [Google Scholar]
- 19.Abramoff, M. D., Magelhaes, P. J., and Ram, S. J. (2004) Biophotonics Int. 11 36-42 [Google Scholar]
- 20.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 21.Lee, H. C., Inoue, T., Imae, R., Kono, N., Shirae, S., Matsuda, S., Gengyo-Ando, K., Mitani, S., and Arai, H. (2007) Mol. Biol. Cell 19 1174-1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001) J. Mol. Biol. 305 567-580 [DOI] [PubMed] [Google Scholar]
- 23.Sonnhammer, E. L., von Heijne, G., and Krogh, A. (1998) Proc. Int. Conf. Intell. Syst. Mol. Biol. 6 175-182 [PubMed] [Google Scholar]
- 24.Yang, Y., Cao, J., and Shi, Y. (2004) J. Biol. Chem. 279 55866-55874 [DOI] [PubMed] [Google Scholar]
- 25.Lands, W. E. (1960) J. Biol. Chem. 235 2233-2237 [PubMed] [Google Scholar]
- 26.Ghosh, M., Tucker, D. E., Burchett, S. A., and Leslie, C. C. (2006) Prog. Lipid Res. 45 487-510 [DOI] [PubMed] [Google Scholar]
- 27.Cao, J., Shan, D., Revett, T., Li, D., Wu, L., Liu, W., Tobin, J. F., and Gimeno, R. E. (2008) J. Biol. Chem. 283 19049-19057 [DOI] [PubMed] [Google Scholar]
- 28.Oelkers, P., Behari, A., Cromley, D., Billheimer, J. T., and Sturley, S. L. (1998) J. Biol. Chem. 273 26765-26771 [DOI] [PubMed] [Google Scholar]
- 29.Oelkers, P., Cromley, D., Padamsee, M., Billheimer, J. T., and Sturley, S. L. (2002) J. Biol. Chem. 277 8877-8881 [DOI] [PubMed] [Google Scholar]
- 30.Bosson, R., Jaquenoud, M., and Conzelmann, A. (2006) Mol. Biol. Cell 17 2636-2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamaki, H., Shimada, A., Ito, Y., Ohya, M., Takase, J., Miyashita, M., Miyagawa, H., Nozaki, H., Nakayama, R., and Kumagai, H. (2007) J. Biol. Chem. 282 34288-34298 [DOI] [PubMed] [Google Scholar]
- 32.Hodges, E., Redelius, J. S., Wu, W., and Hoog, C. (2005) Mol. Cell. Proteomics 4 1319-1327 [DOI] [PubMed] [Google Scholar]
- 33.Stahl, U., Stalberg, K., Stymne, S., and Ronne, H. (2007) FEBS Lett. 582 305-309 [DOI] [PubMed] [Google Scholar]
- 34.Sumper, M., and Trauble, H. (1973) FEBS Lett. 30 29-34 [DOI] [PubMed] [Google Scholar]