Abstract
Mutations in p97/VCP cause the multisystem disease inclusion body myopathy, Paget disease of the bone and frontotemporal dementia (IBMPFD). p97/VCP is a member of the AAA+ (ATPase associated with a variety of activities) protein family and has been implicated in multiple cellular processes. One pathologic feature in IBMPFD is ubiquitinated inclusions, suggesting that mutations in p97/VCP may affect protein degradation. The present study shows that IBMPFD mutant expression increases ubiquitinated proteins and susceptibility to proteasome inhibition. Co-expression of an aggregate prone protein such as expanded polyglutamine in IBMPFD mutant cells results in an increase in aggregated protein that localizes to small inclusions instead of a single perinuclear aggresome. These small inclusions fail to co-localize with autophagic machinery. IBMPFD mutants avidly bind to these small inclusions and may not allow them to traffic to an aggresome. This is rescued by HDAC6, a p97/VCP-binding protein that facilitates the autophagic degradation of protein aggregates. Expression of HDAC6 improves aggresome formation and protects IBMPFD mutant cells from polyglutamine-induced cell death. Our study emphasizes the importance of protein aggregate trafficking to inclusion bodies in degenerative diseases and the therapeutic benefit of inclusion body formation.
Ubiquitinated inclusions (UBIs)2 are a common pathology in many degenerative disorders associated with brain and muscle (1). In many cases the molecular constituents of the UBIs are known and are a result of the misfolding of an aggregate prone or mutant protein such as α-synuclein in autosomal dominant Parkinson disease or expanded polyglutamine containing proteins in Huntington disease. UBIs in other degenerative disorders may be due to a global impairment in protein degradation. For example, after a misfolded substrate overwhelms the ubiquitin proteasome system (UPS) or as a result of mutations in proteins involved in specific protein degradation pathways. One example is the autosomal dominantly inherited disorder inclusion body myopathy, Paget disease of the bone and frontotemporal dementia (IBMPFD) associated with mutations in the UPS chaperone p97/VCP (2). Affected tissue in IBMPFD contains prominent cytoplasmic and intranuclear UBIs (3-5). In some cases these UBIs contain tubulofilamentous inclusions and insoluble protein aggregates (4). p97/VCP belongs the AAA+ (ATPases associated with a variety of activities) protein family and participates in the degradation of proteins via the UPS (6). p97/VCP has a clear role in endoplasmic reticulum-associated degradation (ERAD) of misfolded proteins (6). In association with co-factors derlin-1, Ufd1, and Npl4, p97/VCP participates in the retrotranslocation of misfolded endoplasmic reticulum lumen and transmembrane proteins facilitating their delivery to the UPS. We have previously shown that IBMPFD mutant p97/VCP fails to degrade the misfolded ERAD substrate ΔF508-CFTR as efficiently as wild type p97/VCP in cultured myoblasts (7). p97/VCP may also facilitate the degradation of soluble cytosolic proteins. Recently, IBMPFD mutations were found to impair the degradation of the cytosolic protein, Unc-45 (8). Whether IBMPFD mutations in p97/VCP affect the degradation of all UPS substrates resulting in UBIs is unknown.
UBIs can also form in the setting of impaired macroautophagy, herein referred to as autophagy (9). Autophagy is a nonselective mechanism for degrading long-lived proteins and organelles, whereas the UPS is a regulated means of targeting short-lived proteins to the proteasome. Genetic inactivation of autophagy in mouse central nervous system tissue results in prominent UBIs and neurodegeneration (10). Autophagy and the UPS have traditionally been thought to serve complementary yet parallel functions in protein homeostasis, and the inter-relationship between these two degradation systems is unclear (11). One potential point of intersection between autophagy and the UPS is the “aggresome” or inclusion body (12). An aggresome is a microtubule-dependent pericentriolar region of the cell that contains sequestered misfolded or aggregated proteins (13). Aggresome formation occurs in the setting of UPS dysfunction because of decreased proteasome activity or the overwhelming accumulation of misfolded proteins (13). The aggresome also contains proteins such as LC3 and p62 along with lysosomes, suggesting that these are areas of active autophagic degradation (14, 15). Ubiquitinated and aggregated proteins are trafficked to the aggresome via interactions with HDAC6 and dynein (13, 15, 16). The molecular machinery involved in triaging degradation destined proteins to the UPS or autophagic pathways remains to be elucidated. This putative “segregase” would need to interact with ubiquitinated substrates as well as with both UPS and autophagic machinery.
The current study examines the role of p97/VCP and its mutants in inclusion body formation and protein aggregate clearance. Although it is clear that p97/VCP plays a critical role in the ERAD degradation of misfolded proteins (6), the role of p97/VCP in the degradation of cytosolically derived misfolded and aggregated proteins is less clear. Loss of p97/VCP function in mammalian cells leads to the accumulation of insoluble ubiquitinated proteins (17-19), similarly IBMPFD mutant p97/VCP expression in myoblasts and transgenic mouse muscle leads to the accumulation of UBIs (7, 20). p97/VCP may be involved in inclusion body formation. Loss of p97/VCP or expression of a dominant negative ATP hydrolysis-deficient mutant impairs aggresome formation (19, 21, 22). p97/VCP associates with the aggresome essential protein, HDAC6 (23, 24). Overexpression of HDAC6 facilitates the degradation of expanded polyglutamine containing androgen receptor or ubiquitinated proteins in an autophagy-specific manner (25). We propose that p97/VCP associates with aggregated proteins and triages them to an inclusion body via interactions with HDAC6. IBMPFD mutants fail to release aggregated proteins, resulting in failed inclusion body formation and protein aggregate clearance.
EXPERIMENTAL PROCEDURES
Plasmid Constructs and Cell Culture—The following plasmids are previously described: pcDNA4.0/TO p97/VCP-His/Myc and pcDNA3.1p97/VCP-Myc/His (17). IBMPFD mutations were introduced into these vectors using site-directed mutagenesis (Stratagene) and sequenced for the integrity of the p97/VCP sequence and verification of their respective point mutations. For lentiviral constructs, a BamHI-AgeI-digested fragment from pcDNA3.1Myc/His vector with the murine p97/VCP-fused to a Myc tag (containing wild type and E578Q, R95G, R155H, and A232E point mutations) was cloned into the BamHI-AgeI site of CCIV FM1 (lentiviral shuttle vector with a cytomegalovirus promoter) to generate the lentiviral constructs p97/VCP-Myc-IRES-VenusGFP. Lentiviruses are prepared as previously described (26). Lentiviral particles released into tissue culture serum are titered by counting the number of GFP fluorescing cells following transduction into HEK293 cells. polyQ80-CFP was created from the parent vector polyQ80-GFP obtained from Dr. Gene Johnson (Washington University). Expression constructs containing p97/VCP-DsRed-WT, E305Q/E578Q, R155H, R95G, and A232E were a gift from Dr. Nigel Cairns (Washington University), pCMV-GFP-LC3 was gift from Dr. Robert Baloh (Washington University), and mcherry-ATG5 and HDAC6-FLAG constructs were obtained via Addgene. pcDNA4.0/TO-polyQ80-CFP was generated by digesting the polyQ80-CFP containing plasmid with BamHI-NotI and ligating to the BglII-NotI sites in pcDNA4.0/TO.
U2OS TRex cells stably transfected with a tetracycline repressor plasmid (Invitrogen) were transfected with pcDNA4.0/TO vectors to generate cell lines expressing p97/VCP. Medium containing 50 μg/ml hygromycin B (Invitrogen) and 125 μg/ml zeocin (Invitrogen) was added to select the stable cell lines. Approximately 12 days later, colonies of zeocin-resistant cells appeared. On average 25-30 colonies for each desired cell line were transferred into separate wells and screened. Immunofluorescence and Western blot analysis were used to identify lines that expressed protein only in the presence of tetracycline. The cells were maintained with 50 μg/ml hygromycin B and 65 μg/ml zeocin, with the exception of nonstably transfected U20S cells, which were maintained with hygromycin B. Protein expression was induced by the addition of 1 μg/ml tetracyline for the indicated times. The cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics at 37 °C with 5% CO2 and seeded in appropriate plates and grown to ∼80% confluence on the day of transfection. Transfections were performed with Lipofectamine 2000 (Invitrogen). For stable cell lines, tetracycline was added 16 h prior to plasmid transfection.
Confocal and Electron Microscopy—U20S cells were plated on glass coverslips and transiently transfected with plasmid DNA or induced with tetracycline as above. 24-48 h post-transfection, the cells were washed twice with PBS, fixed with 3% paraformaldehyde in PBS for 15 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, washed, and then blocked with 3% goat serum for 1 h. After primary antibody incubation, the cells were incubated with Alexa Fluor-conjugated secondary antibody for 1 h, washed several times, and mounted in antifade solution. The samples were observed using a confocal microscope (Carl Zeiss, Jena, Germany).
For thin section electron microscopy, transiently transfected cells were grown to 70% confluence in a 100-mm dish for 36 h. For sample preparation, trypsinized cells were collected, washed in phosphate-buffered saline, fixed in 2.5% glutaraldehyde in sodium cacodylate, embedded, sectioned, and stained with uranyl acetate according to standard procedures.
Fluorescence Recovery after Photobleaching—The cells were plated and transfected 36 h prior to imaging on glass-bottomed dishes. Immediately before imaging, the medium was replaced with medium containing 10 mm Hepes buffer, pH 7.5. Imaging and photobleaching were performed using a Plan NEOFLUAR 63×/1.30 oil objective on an inverted confocal microscope (model LSM510 Meta; Carl Zeiss MicroImaging, Inc.). The cells transfected with CFP fusion proteins were imaged with 488-nm light and DsRed with 516-nm light, using 2% laser power and a pin hole of 1 airy unit. After two imaging scans, a selected area of the inclusion (region of interest) was bleached using maximal laser power for 10 iterations, and then the photobleached cell was imaged at 20-s intervals for 10 min. The collected images were analyzed using Zeiss LSM 5 software to calculate the mean fluorescence intensity in the region of interest as a function of time after photobleaching. The relative fluorescence intensity values are corrected by percentage of prebleaching values.
Quantification of Inclusions—Normal U2OS cells and U2OS cells stably expressing tetracycline-inducible p97/VCP-WT or IBMPFD mutants were induced with tetracycline and transfected with polyQ80-CFP for 48 h and then processed for fluorescence microscopic analysis of inclusion formation. The cells were counted. For inclusions, 10 random fields of each sample were selected, and the cells containing inclusions were manually counted for the number of inclusions/cell. ∼100 cells/experiment were counted and averaged among three replicates totaling ∼300 cells/condition. Counted cells were grouped into two categories: ≤3 inclusions/cell or >3 inclusions/cell, and the data were subjected to statistical analysis. Inclusion clearance was performed in a similar manner, except that total transfected cells were counted and the percentage of inclusion containing cells/total transfected cells were plotted for each time point and condition.
Filter Trap Assay—Normal U2OS cells and U2OS cells stably expressing tetracycline-inducible p97/VCP-WT or IBMPFD mutants were induced and transfected with polyQ80-CFP for 48 h. The cells were harvested using 1× PBS including protease inhibitors and brief sonication. Twenty μg of protein were mixed with 900 μl of PBS + 1% SDS. Subsequently, the samples were applied to a slot blot unit (Bio-Rad, Bio-Dot) and filtered through a nitrocellulose membrane. The membrane was probed with anti-GFP antibody.
Immunoprecipitation and Western Blotting—Immunoprecipitation was performed using Seize Classic(G) immunoprecipitation kit (Pierce). The cells were washed with cold PBS, scraped, pelleted by centrifugation, and lysed on ice for 30 min with radioimmune precipitation assay buffer containing protease inhibitors. The cell lysate were briefly sonicated, centrifuged for 10 min at 12,000 × g at 4 °C, and the supernatants were used for immunoprecipitation. One hundred microliters of cell lysate was incubated with 2 μg of mouse monoclonal anti-Myc antibody. After overnight incubation at 4 °C with rotation, immune complex, and 400 μl of protein G (50% slurry) beads were placed into a spin cup column and incubated for 2 h at room temperature. The bound proteins were eluted from the beads, boiled for 5 min, and analyzed by Western blotting. Western blot analysis was performed as described previously (7). The following antibodies were used: anti-p97 (BD Biosciences), anti-LC3 (NanoTools), anti-p62 (Abgent), anti-HDAC6, anti-Myc, and anti-tubulin (Cell Signaling Technology), anti-actin (sigma), anti-FK2 (Biomol), and rabbit polyclonal anti-GFP (B5).
Electroporation into Mouse Tibialis Anterior (TA)—3-month-old female mice were anesthesized with pentobarbital, and their hindlimb was shaved. 20 μg of polyQ80-CFP expression construct resuspended in 50 μl of 1× PBS was injected into the TA with a 27-gauge needle. A 5-mm gap BTX 2 needle array attached to a BTX ECM 830 electroporator was set for 100 V, pulse length of 50 ms, pulse interval of 200 ms, and six pulses. The animals were allowed to recover, and TA muscle was harvested 7 days later.
MTT/MTS Assays—U2OS stable cells were seeded in 6-well plates and allowed to attach overnight, and then the cells were induced. Transfection with 1.5 μg of polyQ80-CFP or polyQ19-CFP DNA was carried out using Lipofectamine 2000 (Invitrogen). Twelve hours post-transfection, the cells were harvested and replated into 96-well plates. A CellTiter 96™ AQueous nonradioactive cell proliferation assay (Promega) kit containing the tetrazolium compound MTS was used according to the manufacturer's instructions to measure the number of cells. MTS color change was monitored by using an ELX-800 universal plate reader (Bio-Tek, Winooski, VT) set at an absorbance reading of 490 nm. For proteasome sensitivity assays, the cells were plated in 96-well plates and treated with vehicle or 10-fold dilutions of 1 mm MG132 or lactacystin. MTS assays were performed 16 h later, and the ratio of cells was determined via comparison with vehicle-treated controls for each.
Statistical Analysis—The data were evaluated by analysis of variance followed by Fishers LSD post hoc comparisons at p < 0.05.
RESULTS
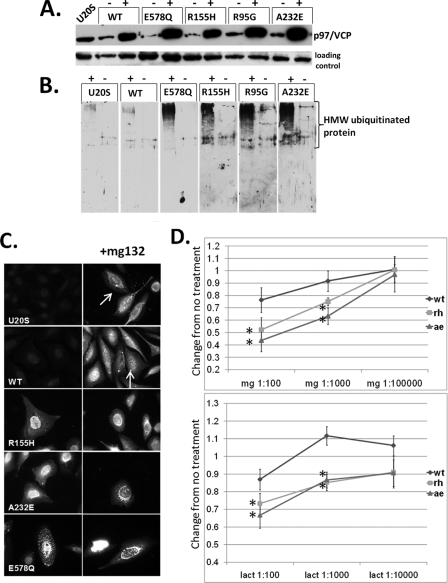
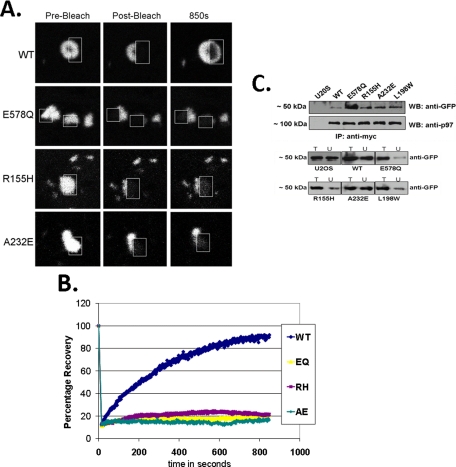
IBMPFD Mutant p97/VCP Impairs Inclusion Body Formation—We generated U20S cell lines that stably express tetracycline-inducible p97/VCP-WT, non-disease-associated ATPase inactive p97/VCP-E578Q or one of four different IBMPFD mutants (R155H (RH), R95G (RG), A232E (AE), and L198W (LW)) with a C-terminal Myc tag (Fig. 1A). Consistent with our previous studies (7), IBMPFD mutant p97/VCP and ATPase inactive p97/VCP-E578Q expression resulted in an increase in high molecular weight ubiquitinated proteins as seen via immunoblot or immunofluorescence 16 h after tetracycline induction (Fig. 1, B and C). When these cells were stressed with sublethal doses of the proteasome inhibitors MG132 or lactacystin, IBMPFD-expressing cells had fewer cells following treatment consistent with enhanced cell death as measured via MTT/MTS assay (Fig. 1D). Immunohistochemistry of MG132-treated cells with an anti-ubiquitin antibody (FK2) showed that only control U20S and p97/VCP-WT-expressing cells generated small perinuclear ubiquitin-positive inclusions consistent with an aggresome (Fig. 1C). In contrast, IBMPFD mutant-expressing cells had a perinuclear increase in ubiquitinated proteins but no clear aggresome (Fig. 1C). p97/VCP-E578Q-expressing cells had an increase in punctate endoplasmic reticulum-associated ubiquitinated inclusions as previously reported (17). This finding suggests that IBMPFD mutant cells have impaired ubiquitinated protein aggregate handling to inclusion bodies, resulting in increased sensitivity to proteasome inhibition. Alternatively, the absence of ubiquitin-positive perinuclear inclusions may more generally reflect an overwhelming of the UPS system in IBMPFD mutant-expressing cells.
FIGURE 1.
IBMPFD mutant-expressing cells increase ubiquitinated proteins. A, characterization of tetracycline-inducible U20S cell lines expressing Myc-tagged p97/VCP, ATPase inactive p97/VCP-E578Qm, or IBMPFD mutant p97/VCP-R155H, R95G, or A232E. Cell lysates from uninduced (-) and 16 h tetracycline-induced (+) U20S cells were separated by SDS-PAGE and subjected to Western blot analysis to detect Myc-tagged p97/VCP (top panel) or actin (bottom panel). B, the same induced cells as above were immunoblotted with an antibody to ubiquitin. The lanes were taken from the same gel at the same exposure and realigned for presentation purposes. Note the increase in high molecular weight ubiquitinated species in IBMPFD mutant-expressing cells. C, ubiquitin immunostaining of control U20S cells or U20S cells expressing p97/VCP-WT, ATPase inactive p97/VCP-E578Q, or IBMPFD mutant p97/VCP-R155H or -A232E either untreated (left panels) or treated with proteasome inhibitor MG132 (right panels). Note that p97/VCP-WT-expressing cells or untransfected cells have small perinuclear aggresomes that are not present in IBMPFD mutant-expressing cells. D, MTT assays of U20S cells expressing p97/VCP-WT or IBMPFD mutant p97/VCP-R155H or -A232E following 16 h of application of 10-fold dilutions (original concentrations, 1 mm) as indicated of the proteasome inhibitors MG132 (mg, top panel) or lactacystin (lact, bottom panel). Units are the change of treated cells compared with untreated cells (arbitrarily set to 1) in each condition.
To see whether IBMPFD mutants mishandled aggregated proteins without exogenous proteasome inhibitors, we co-expressed an expanded polyglutamine containing fusion protein (polyQ80-CFP) in U20S cells. PolyQ80-CFP intrinsically aggregates and forms inclusion bodies consistent with an aggresome. Aggresomes are a cellular response to increased levels of misfolded or aggregated proteins (13). They are localized to the microtubule organizing center and are surrounded by an intermediate filament network (13). Although aggresomes contain aggregated protein, they are distinct from other protein aggregates because they are formed via an active sequestration of misfolded and aggregated proteins that requires an intact microtubule network. Therefore a protein or protein aggregate that fails to be rapidly degraded via the UPS will coalesce into these larger inclusion bodies or aggresomes (13). Once in the aggresome, undegraded ubiquitinated and aggregated proteins are thought to be degraded via autophagy (27).
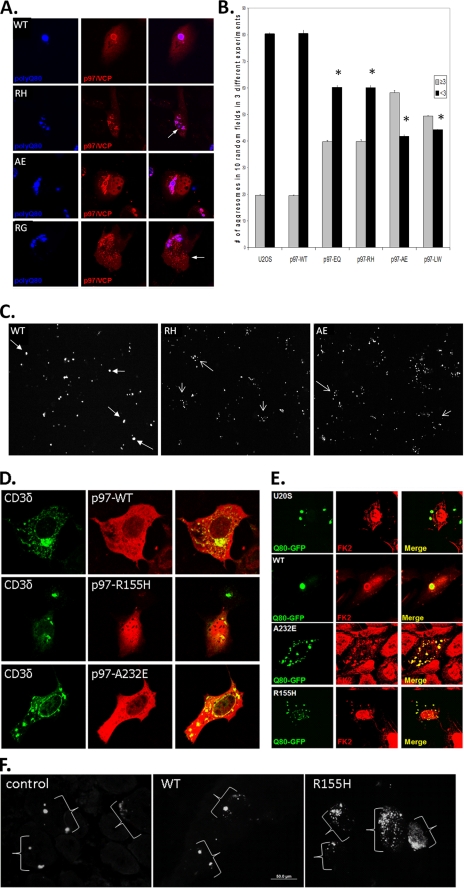
Following co-expression of polyQ80-CFP in cells expressing p97/VCP-WT fused to a DSred fluorescent tag, we saw p97/VCP-WT surrounding the inclusion body. Inclusion bodies in p97/VCP-WT-expressing cells were predominantly single or <3 inclusions/transfected cell (Fig. 2A) and had properties consistent with an aggresome (not shown). In contrast, there was a decrease in single perinuclear polyQ80-CFP inclusion bodies (≤3 inclusions/cell) in IBMPFD mutant-expressing cells. Instead there was an increase in smaller cytosolic inclusions (>3 inclusions/cell) that did not always co-localize with the microtubule organizing center. p97/VCP-R155H, R95G, and A232E proteins co-localized with polyQ80-CFP, although there were both polyQ80-CFP and p97/VCP-IBMPFD inclusions that did not co-localize (Fig. 2A, see arrows). An aggresome is traditionally defined as a single perinuclear inclusion body (13); however, for purposes of quantitation, we counted the number of cells with ≤3 inclusions (aggresomal) or >3 inclusions (non-aggresomal). These data are graphically represented in Fig. 2B and by a low magnification field of p97/VCP-WT or IBMPFD mutant-expressing cells 48 h after transfection with polyQ80-CFP (Fig. 2C). Impaired inclusion body or aggresome formation was seen following expression of other proteins known to accumulate in aggresomes such as ΔF508-CFTR (7) and CD3δ-YFP (Fig. 2D). PolyQ80 inclusions in both p97/VCP-WT and IBMPFD mutant p97/VCP co-localized with ubiquitin (Fig. 2E). These data suggest that IBMPFD mutants mishandle ubiquitinated and aggregated proteins by failing to send them to a single inclusion body.
FIGURE 2.
Aggresome formation is impaired in IBMPFD mutant-expressing cells and tissue. A, representative live cell images of cells co-expressing polyQ80-CFP (blue) with DsRed tagged p97/VCP-WT (WT) or IBMPFD mutants R155H (RH), A232E (AE), and R95G (RG). Note that although p97/VCP co-localizes with polyQ80-CFP, there is impaired inclusion formation in IBMPFD mutant-expressing cells. B, control U20S or U20S cells stably expressing tetracycline-inducible p97/VCP-WT, ATPase inactive p97/VCP-E578Q (EQ) or IBMPFD mutant p97/VCP R155H, A232E, and L198W (LW) were transfected with polyQ80-CFP for 48 h. The cells were fixed with 3% paraformaldyhyde and processed for fluorescence microscopic analysis of inclusion bodies. The cells were scored for the presence of ≤ 3 inclusions or >3 inclusions. *, p < 0.05 versus U20S control containing less than three aggregated proteins. C, representative low power fields from U20S cells stably expressing tetracycline-inducible p97/VCP-WT or IBMPFD mutant p97/VCP R155H and A232E transfected with polyQ80-CFP for 48 h. The filled arrows denote cells with ≤3 inclusions, and the open arrows denote cells with >3 inclusions. D, confocal micrograph of p97/VCP-dsRed and CD3δ-YFP co-expression. Note that overexpressed CD3δ-YFP forms a perinuclear inclusion in p97/VCP-WT-expressing cells and generates multiple smaller inclusions in IBMPFD mutant cells (R155H and A232E). E, control or U20S cells expressing p97/VCP-WT or IBMPFD mutant p97/VCP R155H or A232E along with polyQ80-CFP. Ubiquitinated proteins were identified via immunohistochemical analysis with FK2 antibody. Note that polyQ80-CFP inclusions are ubiquitin-positive. F, non-transgenic control, p97/VCP-WT or p97/VCP-R155H-expressing transgenic animals electroporated with a polyQ80-CFP expression construct into the TA muscle 7 days prior to analysis. Images are CFP autofluorescence from the TA muscles. Control and p97/VCP-WT skeletal muscle typically have a few large inclusion bodies/myofiber, although IBMPFD mutant R155H transgenic muscle has multiple small inclusion bodies occasionally surrounding a larger inclusion body. The brackets denote groups of inclusions within single myofibers.
To see whether impaired aggregate handling to an inclusion body occurred in vivo, we electroporated a polyQ80-CFP expression construct into the tibialis anterior of control, p97/VCP-WT, or one of two independent IBMPFD mutant p97/VCP-R155H-expressing transgenic mouse lines (20). After 7 days of expression, the animals were sacrificed and analyzed for polyQ80-CFP fluorescence. Similar to our cell culture studies, polyQ80-CFP formed large inclusions in transfected myofibers from control or p97/VCP-WT-expressing mice (Fig. 2F). In contrast, IBMPFD mutant R155H-expressing animals had many fibers with multiple small inclusions that failed to coalesce into a larger inclusion body (Fig. 2F). These data suggest that protein aggregates coalesce less reliably into a single inclusion body in IBMPFD mutant-expressing skeletal muscle as well.
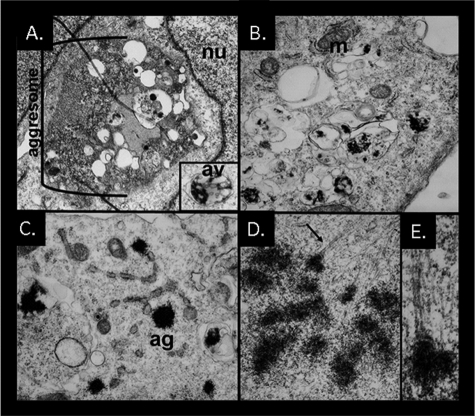
To define the localization of the polyQ80-CFP inclusions, we performed transmission electron microscopy on cells co-expressing p97/VCP-WT or IBMPFD mutants (p97/VCP-R155H or A232E) along with polyQ80-CFP. p97/VCP-WT-expressing cells had large perinuclear inclusions or aggresomes containing filamentous material (Fig. 3A). These inclusions typically had membranous vacuoles with single and double membranes. Many vacuoles contained electron dense filamentous debris and were characteristic of autophagosomes (Fig. 3A, inset). In contrast, IBMPFD mutant-expressing cells rarely had a large single perinuclear inclusion but instead had smaller electron dense inclusions both within vacuolar structures or within the cytoplasm admixed around organelles (Fig. 3, B and C). In some cases electron-dense inclusions appeared stuck at the ends of microtubules (Fig. 3, D and E). These data further support that polyQ80 aggregates fail to traffic to an aggresome in IBMPFD mutant-expressing cells.
FIGURE 3.
Aggresome formation is impaired in IBMPFD mutant-expressing cells and tissue. Electron micrographs of U20S cells co-expressing p97/VCP-WT (A) or p97/VCP-R155H (B-E) along with polyQ80-CFP for 48 h. A, p97/VCP-WT-expressing cells have large perinuclear inclusions with active autophagic degradation consistent with an aggresome. The inset shows a double membrane containing autophagic vacuole with electron dense debris. B and C, IBMPFD mutant-expressing cells have multiple small electron dense inclusions that are within membranous structures or non-membrane-associated within the cytoplasm. D and E, an IBMPFD mutant-expressing cell with multiple inclusions attached to microtubules. Similar results were also obtained with p97/VCP-A232E-expressing cells.
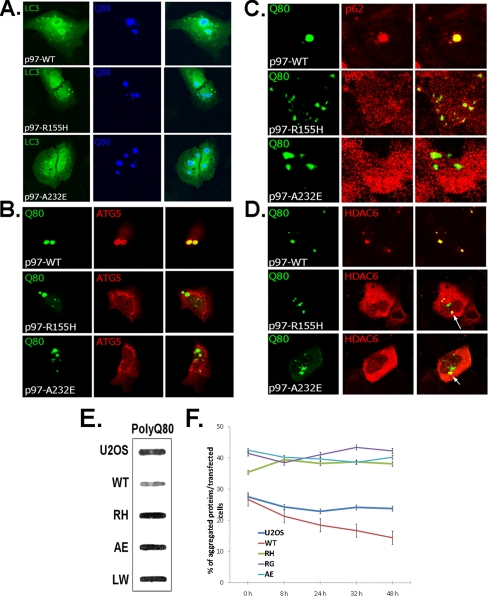
IBMPFD Mutant p97/VCP Impairs the Degradation of Protein Aggregates—It has been suggested that protein aggregates are trafficked to a perinuclear inclusion body as a means of facilitating their autophagic degradation (27). We evaluated whether the protein aggregates seen in our IBMPFD mutant-expressing cells co-localized with autophagic proteins. In agreement with our ultrastructural data, co-expressed GFP-LC3, a marker for autophagosomes, fails to co-localize as robustly with many polyQ80-CFP inclusions in IBMPFD mutant-expressing cells (Fig. 4A). This was in contrast to p97/VCP-WT-expressing cells, which had consistent co-localization to GFP-LC3. Similar results were seen following co-transfection with mCherry-ATG5 (Fig. 4B) or immunostaining with an antibody to p62 (Fig. 4C) and HDAC6 (Fig. 4D). There was not a complete absence of LC3, ATG5, and p62-positive inclusions in IBMPFD mutant cells. In fact in the case of ATG5 and p62, there was a significant increase in immunoreactivity that did not co-localize with polyQ80-CFP (Fig. 4, B and C). Furthermore in the case of HDAC6, whereas every polyQ80-CFP inclusion co-localized with HDAC6 in p97/VCP-WT cells, the pattern was much more diffuse, and usually one of the multiple inclusions co-localized with HDAC6 in IBMPFD mutant-expressing cells (Fig. 4D, see arrow). In IBMPFD mutant-expressing cells that formed a single perinuclear aggresome, co-localization with autophagic markers was more evident. These data suggest IBMPFD mutant-expressing cells fail to send protein aggregates to a single perinuclear inclusion body, not allowing these aggregates to co-localize with protein degradation machinery.
FIGURE 4.
Polyglutamine inclusions fail to co-localize and are less efficiently degraded by the autophagic machinery in IBMPFD mutant-expressing cells. U20S cells stably expressing tetracycline-inducible p97/VCP-WT or IBMPFD mutant p97/VCP R155H, A232E were co-transfected with polyQ80-CFP and expression vectors GFP-LC3 (A) and mcherry-ATG5 (B) or immunostained to visualize endogeneous p62 (C) and HDAC6 (D). Representative confocal images are shown. All of the images were taken at the same gain and laser power, allowing comparison in the intensity of fluorescence between samples. E, control or U20S cells stably expressing tetracycline-inducible p97/VCP-WT or IBMPFD mutant p97/VCP R155H, A232E, and L198W were transfected with polyQ80-CFP. 48 h later lysates were prepared in 1% SDS and filtered through a nitrocellulose membrane and blotted with anti-GFP antibody. Note the darker band in IBMPFD mutant-expressing lysates. Total polyQ80 protein levels were similar between cell lines (see C). F, U20S cells expressing a tetracycline-inducible polyQ80-CFP were transfected with p97/VCP-WT or IBMPFD mutant p97/VCP R155H, R95G, and A232E. Forty-eight hours after transfection, the cells were washed with PBS to remove tetracycline and incubated in medium without tetracycline. Aggregate-containing transfected cells were counted and plotted as the percentages of aggregates/total transfectants.
Because polyQ80-CFP aggregates fail to form perinuclear aggresomes and co-localize with autophagic machinery, we evaluated the degradation of polyQ80-CFP. Co-expression of IBMPFD mutant p97/VCP for 48 h with polyQ80-CFP resulted in an increase in aggregated polyQ80-CFP as demonstrated by filter trap (Fig. 4E). p97/VCP-WT co-expression had less aggregated polyQ80-CFP than non-p97/VCP co-transfected cells (U20S), suggesting that p97/VCP-WT may facilitate the degradation of protein aggregates (Fig. 4E). When this experiment was performed with a nonaggregating polyglutamine (polyQ19-CFP) or GFP alone, there was no increase in the aggregated GFP or polyQ19-CFP in IBMPFD mutant-expressing cells (not shown). To confirm that the degradation of polyQ80 was impaired, resulting in the increase in aggregated protein, we used U20S cells that expressed a tetracycline-inducible polyQ80-CFP plasmid construct. This construct expresses polyQ80-CFP in the presence of tetracycline and fails to express polyQ80-CFP in the absence of tetracycline. 36 h after induction with tetracycline, polyQ-CFP aggregates were present in ∼30% of the cells (Fig. 4F; time, 0 h). When a constitutively expressed IBMPFD mutant p97/VCP-Myc was co-expressed with polyQ-CFP, there was an increase in the number of transfected cells with CFP-positive aggregates to ∼40% (Fig. 4F; time, 0 h). Following tetracycline washout, there was a decrease in the number of CFP-positive aggregates/transfected cells in non-p97/VCP overexpressing or p97/VCP-WT-Myc-expressing cells as early as 8 h that continued for 48 h (Fig. 4F). This was in contrast to IBMPFD mutant p97/VCP-expressing cells that failed to clear the polyQ80-CFP aggregates (Fig. 4F). These data demonstrate that the clearance of protein aggregates is impaired in IBMPFD mutant-expressing cells.
IBMPFD Mutant p97/VCP “Traps” Aggregated Polyglutamine—Given that IBMPFD mutant p97/VCP co-localized with polyQ80-CFP in the absence of a single perinuclear inclusion, we reasoned that IBMPFD mutant p97/VCP may be “holding” the polyQ80 protein and not allowing it to traffic to the aggresome. To test this, we performed fluorescence recovery after photobleaching (FRAP) on cells expressing p97/VCP-DsRed and polyQ80-CFP. We photobleached one-half of a polyQ80-CFP p97/VCP-positive inclusion and imaged p97/VCP-DsRed for 15 min to evaluate the live cell dynamics of p97/VCP cycling on and off of aggregated substrate (polyQ80) in the cell. Photobleaching of p97/VCP-WT with associated polyQ80 inclusions resulted in recovery of the transient bleach zone, whereas photobleaching of a dominant negative p97/VCP-E305Q/E578Q (EQ) (that lacks the ability to hydrolyze ATP) associated with a polyQ80 inclusion led to minimal p97/VCP fluorescence recovery (Fig. 5, A and B). This was similar to IBMPFD mutant p97/VCP mutant R155H (RH) and A232E (AE) associated polyQ80 inclusions, which also fail to recover after photobleaching (Fig. 5, A and B). These results suggest that IBMPFD mutant p97/VCP may effectively “hold or trap” aggregated proteins and not allow them to move to an aggresome and associate with autophagic machinery.
FIGURE 5.
IBMPFD mutants are “trapped” on aggregated polyglutamine. A, FRAP analysis of the molecular interaction between polyQ80-CFP and p97/VCP-DsReds (WT, E305Q/E578Q, R155H, or A232E) co-localizing inclusions. The relative fluorescence intensity was determined for each time point and is represented as the percentage of the prebleaching relative fluorescence intensity value. FRAP analysis was typically performed on >2 cells/field and >3 fields/experimental condition. Each experiment was repeated three times with different transfectants for each experiment. B, representative FRAP analysis images of co-localized p97/VCP-DsRed/polyQ80-CFP inclusions before bleaching, immediately after photobleaching and after recovery. Inclusions within cells were photobleached in a small region of interest (outlined box) and monitored for recovery of fluorescence into the region of interest. C, U20S control and U20S cells stably expressing tetracycline-inducible p97/VCP-WT-Myc, dominant negative p97/VCP-E578Q-Myc or IBMPFD mutant p97/VCP-R155H-Myc, -A232E-Myc, and -L198W-Myc were transfected with polyQ80-CFP. 36 h later lysates were collected, and p97/VCP was immunoprecipitated with an anti-Myc. The immunoprecipitate was subsequently immunoblotted for associated polyQ80-CFP with anti-GFP. Similar amounts of p97/VCP were immunoprecipitated. Total lysates (T) and unbound (U) fractions were separated by SDS-PAGE and immunoblotted with anti-GFP antibody. All of the blots are representative of three independent experiments.
Biochemical evidence of this phenomenon is shown with co-immunoprecipitation of polyQ80-CFP with p97/VCP-Myc using a Myc antibody followed by Western blotting with an anti-GFP antibody. A greater amount of polyQ80-CFP was immunoprecipitated with IBMPFD mutant p97/VCP than p97/VCP-WT (Fig. 5C). p97/VCP-E578Q, which lacks the ability to hydrolyze ATP and behaves as a “substrate trap,” serves a positive control and binds a significant amount of polyQ80-CFP (Fig. 5C).
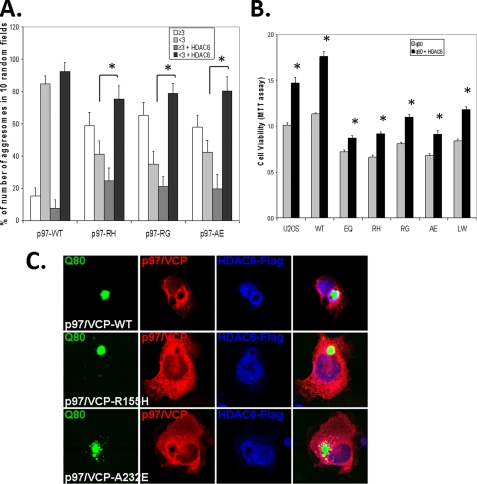
We reasoned that HDAC6, a ubiquitin-binding protein that has been shown to interact with p97/VCP, may be able to facilitate aggresome formation in IBMPFD mutant-expressing cells (24). When we co-expressed HDAC6-FLAG along with polyQ80-CFP and p97/VCP-WT or IBMPFD mutant p97/VCP, HDAC6-FLAG expression significantly increased the number of cells containing ≤3 inclusions in our IBMPFD mutant-expressing cells (Fig. 6A) and co-localized to polyQ80 inclusions (Fig. 6C). In addition, IBMPFD mutant p97/VCP-expressing cells are more sensitive to polyQ80-induced cell death, and HDAC6 co-expression rescued them from polyQ80-induced cell death (Fig. 6B). This was also true for control and p97/VCP-WT-expressing cells.
FIGURE 6.
HDAC6 co-expression improves aggresome formation and rescue cell death in IBMPFD mutant-expressing cells. A, quantification of polyQ80-CFP-positive inclusion bodies (cells containing <3or ≥3 inclusions) in p97/VCP-WT or IBMPFD mutant-expressing (R155H, R95G, and A232E) cells with or without co-expression of HDAC6-FLAG. Note that all IBMPFD mutant cell lines have fewer cells with <3 inclusion than p97/VCP-WT. In addition, HDAC6 increases the number of cells with <3 inclusions in IBMPFD mutant-expressing cells. Non-HDAC6 transfected controls (<3 inclusions/cell) of each group versus HDAC6 transfected (<3 inclusions/cell) of each group have a p < 0.05 (*). B, control U20S cells and U20S cells stably expressing tetracycline-inducible p97/VCP-WT or IBMPFD mutant p97/VCP-R155H, -R95G, and -A232E were transfected with polyQ80-CFP with or without co-expression of HDAC6-FLAG. Forty-eight hours after transfection, the cell number was measured via MTT assay. Note that the number of IBMPFD mutant p97/VCP expressing is decreased compared with p97-VCP-WT, and HDAC6 increases the number of cells when co-expressed with IBMPFD mutant p97/VCP. Cell death is given as a ratio of the nontransfected controls of each group. The results are means ± S.E. and representative of at least three independent experiments. *, p < 0.05 versus non-HDAC6 overexpressing controls of each group. C, representative confocal micrographs of cells expressing p97/VCP-Myc, polyQ80-CFP, and HDAC6-FLAG. Note the co-localization of HDAC6 with a perinuclear inclusion body in IBMPFD mutant-expressing cells.
DISCUSSION
It is well established that p97/VCP is essential for the degradation of some proteins via ERAD and the UPS (6). p97/VCP may also be involved in the “handling” of misfolded and aggregated proteins. p97/VCP associates with pathologic protein inclusions in several diseases. These include Lewy bodies in Parkinson disease, SOD-positive inclusions in amyotrophic lateral sclerosis, and huntingtin-positive inclusions in Huntington disease (28, 29). In addition, p97/VCP is necessary for “aggresome” formation and associates with HDAC6, a ubiquitin-binding protein that shuttles undegraded proteins to the aggresome and facilitates their autophagic degradation (21, 23).
p97/VCP also participates in the degradation of protein aggregates. In particular, overexpression of either of the two Caenorhabditis elegans p97/VCP homologues decreases expanded polyglutamine inclusions in a C. elegans disease model (30). In addition, preformed polyglutamine or ubiquitinated inclusions are degraded less efficiently following small interfering RNA knockdown of p97/VCP (31). Impaired degradation of aggregated polyglutamine did not occur following small interfering RNA knockdown of p97/VCP specific co-factors essential to its function in the UPS such as Ufd1 or Npl4 but only following p97/VCP knockdown, suggesting that that role of p97/VCP in polyglutamine degradation may be independent of its function in the UPS (32).
The current study further elucidates a role for p97/VCP in inclusion body formation and protein aggregate degradation. IBMPFD mutant p97/VCP bound more avidly to aggregated polyglutamine. This resulted in smaller inclusion bodies that did not coalesce into a larger perinuclear inclusion body or aggresome. This could be reversed via co-expression of HDAC6, which improved aggresome formation and rescued cell death. The degradation of preformed protein inclusions occurs predominantly via autophagy. This was also impaired in IBMPFD mutant-expressing cells, leading to an increase in aggregated polyglutamine.
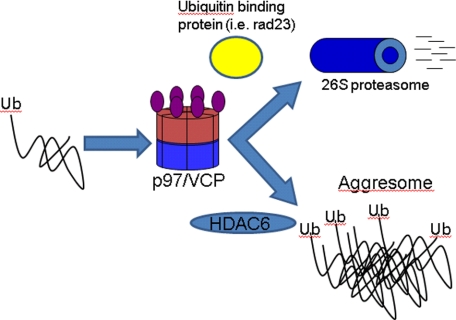
Our results are consistent with existing data. p97/VCP has been shown to associate with HDAC6 (24). Boyault et al. (23) found that increasing levels of HDAC6 favored inclusion body formation, whereas overexpression of p97/VCP favored UPS degradation of ubiquitinated proteins. They proposed a model in which the molar ratios of p97/VCP:HDAC6 dictated the fate of a misfolded protein. Our study found IBMPFD mutants in p97/VCP behave as a substrate trap, similar to ATPase inactive p97/VCP-E578Q and bind ubiquitinated or aggregated proteins more avidly than p97/VCP-WT. In this setting, IBMPFD mutant p97/VCP disrupts the delicate balance between p97/VCP:HDAC6, shifting the equilibrium toward p97/VCP. The consequence is a decrease in single perinuclear inclusion bodies that can be overcome with increasing levels of HDAC6. The increased affinity of the IBMPFD mutants for an aggregated or ubiquitinated protein fails to release it to the UPS as well. This too results in the accumulation of undegraded ubiquitinated proteins. We have previously shown that ΔF508-CFTR, a substrate known to be handled via p97/VCP, has impaired degradation in IBMPFD mutant-expressing cells (7). We suggest that IBMPFD mutants impair both inclusion body formation (and subsequent autophagic clearance) and UPS-mediated degradation. This model places p97/VCP as central fate determiner for aggregated proteins. By means of interactions with HDAC6, p97/VCP sends aggregated proteins to an inclusion body for autophagic clearance. Similarly via interactions with UPS shuttling proteins such as rad23 or the 26 S proteasome itself, p97/VCP facilitates the degradation of misfolded yet soluble proteins (Fig. 7).
FIGURE 7.
Model of proposed p97/VCP function. p97/VCP triages ubiquitinated proteins to the UPS or the aggresome via interactions with other ubiquitin-binding proteins (i.e. rad23) or HDAC6 respectively. IBMPFD mutant p97/VCP avidly binds to ubiquitinated and aggregated proteins failing to efficiently release them to the UPS or aggresome. This results in the accumulation of undegraded ubiquitinated and aggregated proteins.
We previously found that IBMPFD mutant p97/VCP-R155H retains ATPase activity (7). Yet we suggest that IBMPFD mutants behave similar to ATPase hydrolysis deficient p97/VCP-E578Q. The hydrolysis of ATP by p97/VCP confers a large conformational change between the N and D1 domains of p97/VCP (33). Many of the known IBMPFD mutations in p97/VCP reside at the interface between the N and D1 domains. Although IBMPFD mutant R155H retains ATPase activity, a disruption at the N and D1 domain interface may interfere with substrate release, effectively serving as a substrate trap.
We suggest that IBMPFD and perhaps its phenotypic components, inclusion body myopathy, Paget disease of the bone, and frontotemporal dementia with ubiquitinated inclusions, may be disorders of protein aggregate handling and subsequent degradation. For example, the most common genetic cause of Paget disease of the bone is mutations in the ubiquitin-binding domain of p62 (34). p62 is scaffolding protein that associates with polyubiquitinated substrates and LC3 (35). It is present on autophagosomes and degraded via autophagy along with associated ubiquitinated cargo (35). One form of frontotemporal dementia with ubiquitinated inclusions is due to mutations in the multivesicular body biogenesis pathway protein CHMP2B (36). The expression of mutant CHMP2B or the loss of multivesicular body biogenesis leads to impaired autophagic degradation of aggregated proteins (37, 38). Finally, many vacuolar myopathies including Danon disease and Pompe disease are due to mutations in lysosomal proteins (39).
An increase in undegraded and aggregated proteins is detrimental to cells. To contain and facilitate the degradation of protein aggregates that are resistant to proteasomal degradation, cells sequester these aggregates into inclusion bodies or aggresomes (13). Aggresomes are areas of active autophagic degradation and have been proposed to be protective to the cell (27). Although IBMPFD mutant-expressing cells had an increase in protein aggregates, these aggregates were not localized to a single perinuclear aggresome. In addition, these small protein aggregates failed to co-localize with autophagic protein markers such as LC3 and p62 at aggresomes. Our studies suggest that mishandling of protein aggregates and subsequent inability to form an aggresome is detrimental to cells. It suggests that although protein aggregates sequestered in an aggresome are protective, non-aggresome-associated protein aggregates are detrimental to cells. Another protein aggregate disorder that fails to form typical inclusion bodies is Parkin-associated juvenile onset Parkinson disease; this disease lacks typical α-synuclein inclusions or Lewy bodies (40). Parkin is an E3 ligase that has been shown to facilitate Lys-63-linked polyubiquitination of misfolded proteins (41). Lys-63-linked polyubiquitinated proteins are preferentially sent to aggresomes for autophagic degradation via association with HDAC6 and p62 (41, 42). We suggest that mistrafficking of misfolded and aggregated proteins to the aggresome leads to impaired autophagic degradation of protein aggregates and disease. Enhancing interactions with HDAC6 may be a reasonable therapeutic strategy.
This study proposes a role for p97/VCP in the autophagic clearance of protein aggregates. p97/VCP is well positioned to serve as a sensor of protein aggregate state and decide which proteins are degraded via the UPS or autophagy. IBMPFD mutations may impair this function, resulting in the accumulation of ubiquitinated and aggregated proteins sensitizing cells to proteotoxic stressors.
Acknowledgments
We thank Dr. E. Johnson for the polyQ-GFP constructs, Drs. Nigel Cairns and Michael Gitcho for VCP-DsRed fusion constructs, and Drs. Virginia Kimonis and Alan Pestronk for thoughtful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant K08 AG026271 (to C. C. W.). This work was also supported by National Institutes of Health Neuroscience Blueprint Core Grant P30 NS057105 to Washington University. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: UBI, ubiquitinated inclusion; UPS, ubiquitin proteasome system; IBMPFD, inclusion body myopathy, Paget disease of the bone and frontotemporal dementia; ERAD, endoplasmic reticulum-associated degradation; GFP, green fluorescent protein; CFP, cyan fluorescent protein; LSD, least significant difference; WT, wild type; PBS, phosphate-buffered saline; TA, tibialis anterior; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; MTS, tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] inner salt; FRAP, fluorescence recovery after photobleaching.
References
- 1.Schwartz, A. L., and Ciechanover, A. (1999) Annu. Rev. Med. 50 57-74 [DOI] [PubMed] [Google Scholar]
- 2.Watts, G. D., Wymer, J., Kovach, M. J., Mehta, S. G., Mumm, S., Darvish, D., Pestronk, A., Whyte, M. P., and Kimonis, V. E. (2004) Nat. Genet. 36 377-381 [DOI] [PubMed] [Google Scholar]
- 3.Forman, M. S., Mackenzie, I. R., Cairns, N. J., Swanson, E., Boyer, P. J., Drachman, D. A., Jhaveri, B. S., Karlawish, J. H., Pestronk, A., Smith, T. W., Tu, P. H., Watts, G. D., Markesbery, W. R., Smith, C. D., and Kimonis, V. E. (2006) J. Neuropathol. Exp. Neurol. 65 571-581 [DOI] [PubMed] [Google Scholar]
- 4.Hubbers, C. U., Clemen, C. S., Kesper, K., Boddrich, A., Hofmann, A., Kamarainen, O., Tolksdorf, K., Stumpf, M., Reichelt, J., Roth, U., Krause, S., Watts, G., Kimonis, V., Wattjes, M. P., Reimann, J., Thal, D. R., Biermann, K., Evert, B. O., Lochmuller, H., Wanker, E. E., Schoser, B. G., Noegel, A. A., and Schroder, R. (2006) Brain 130 381-393 [DOI] [PubMed] [Google Scholar]
- 5.Schroder, R., Watts, G. D., Mehta, S. G., Evert, B. O., Broich, P., Fliessbach, K., Pauls, K., Hans, V. H., Kimonis, V., and Thal, D. R. (2005) Ann. Neurol. 57 457-461 [DOI] [PubMed] [Google Scholar]
- 6.Halawani, D., and Latterich, M. (2006) Mol. Cell 22 713-717 [DOI] [PubMed] [Google Scholar]
- 7.Weihl, C. C., Dalal, S., Pestronk, A., and Hanson, P. I. (2006) Hum. Mol. Genet. 15 189-199 [DOI] [PubMed] [Google Scholar]
- 8.Janiesch, P. C., Kim, J., Mouysset, J., Barikbin, R., Lochmuller, H., Cassata, G., Krause, S., and Hoppe, T. (2007) Nat. Cell Biol. 9 379-390 [DOI] [PubMed] [Google Scholar]
- 9.Rubinsztein, D. C. (2006) Nature 443 780-786 [DOI] [PubMed] [Google Scholar]
- 10.Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H., and Mizushima, N. (2006) Nature 441 885-889 [DOI] [PubMed] [Google Scholar]
- 11.Ding, W. X., Ni, H. M., Gao, W., Yoshimori, T., Stolz, D. B., Ron, D., and Yin, X. M. (2007) Am. J. Pathol. 171 513-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopito, R. R. (2000) Trends Cell Biol. 10 524-530 [DOI] [PubMed] [Google Scholar]
- 13.Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998) J. Cell Biol. 143 1883-1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjorkoy, G., Lamark, T., Brech, A., Outzen, H., Perander, M., Overvatn, A., Stenmark, H., and Johansen, T. (2005) J. Cell Biol. 171 603-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwata, A., Riley, B. E., Johnston, J. A., and Kopito, R. R. (2005) J. Biol. Chem. 280 40282-40292 [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi, Y., Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A., and Yao, T. P. (2003) Cell 115 727-738 [DOI] [PubMed] [Google Scholar]
- 17.Dalal, S., Rosser, M. F., Cyr, D. M., and Hanson, P. I. (2004) Mol. Biol. Cell 15 637-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojcik, C., Rowicka, M., Kudlicki, A., Nowis, D., McConnell, E., Kujawa, M., and Demartino, G. N. (2006) Mol. Biol. Cell 17 4006-4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojcik, C., Yano, M., and DeMartino, G. N. (2004) J. Cell Sci. 117 281-292 [DOI] [PubMed] [Google Scholar]
- 20.Weihl, C. C., Miller, S. E., Hanson, P. I., and Pestronk, A. (2007) Hum. Mol. Genet. 16 919-928 [DOI] [PubMed] [Google Scholar]
- 21.Kitami, M. I., Kitami, T., Nagahama, M., Tagaya, M., Hori, S., Kakizuka, A., Mizuno, Y., and Hattori, N. (2006) FEBS Lett. 580 474-478 [DOI] [PubMed] [Google Scholar]
- 22.Song, C., Xiao, Z., Nagashima, K., Li, C. C., Lockett, S. J., Dai, R. M., Cho, E. H., Conrads, T. P., Veenstra, T. D., Colburn, N. H., Wang, Q., and Wang, J. M. (2008) Toxicol. Appl. Pharmacol. 228 351-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyault, C., Gilquin, B., Zhang, Y., Rybin, V., Garman, E., Meyer-Klaucke, W., Matthias, P., Muller, C. W., and Khochbin, S. (2006) EMBO J. 25 3357-3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seigneurin-Berny, D., Verdel, A., Curtet, S., Lemercier, C., Garin, J., Rousseaux, S., and Khochbin, S. (2001) Mol. Cell. Biol. 21 8035-8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey, U. B., Nie, Z., Batlevi, Y., McCray, B. A., Ritson, G. P., Nedelsky, N. B., Schwartz, S. L., DiProspero, N. A., Knight, M. A., Schuldiner, O., Padmanabhan, R., Hild, M., Berry, D. L., Garza, D., Hubbert, C. C., Yao, T. P., Baehrecke, E. H., and Taylor, J. P. (2007) Nature 447 859-863 [DOI] [PubMed] [Google Scholar]
- 26.Lee, C. S., Tee, L. Y., Warmke, T., Vinjamoori, A., Cai, A., Fagan, A. M., and Snider, B. J. (2004) J. Neurochem. 91 996-1006 [DOI] [PubMed] [Google Scholar]
- 27.Taylor, J. P., Tanaka, F., Robitschek, J., Sandoval, C. M., Taye, A., Markovic-Plese, S., and Fischbeck, K. H. (2003) Hum. Mol. Genet. 12 749-757 [DOI] [PubMed] [Google Scholar]
- 28.Ishigaki, S., Hishikawa, N., Niwa, J., Iemura, S., Natsume, T., Hori, S., Kakizuka, A., Tanaka, K., and Sobue, G. (2004) J. Biol. Chem. 279 51376-51385 [DOI] [PubMed] [Google Scholar]
- 29.Hirabayashi, M., Inoue, K., Tanaka, K., Nakadate, K., Ohsawa, Y., Kamei, Y., Popiel, A. H., Sinohara, A., Iwamatsu, A., Kimura, Y., Uchiyama, Y., Hori, S., and Kakizuka, A. (2001) Cell Death Differ. 8 977-984 [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka, K., Okubo, Y., Suzaki, T., and Ogura, T. (2004) J. Struct. Biol. 146 242-250 [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, T., Manno, A., and Kakizuka, A. (2007) Genes Cells 12 889-901 [DOI] [PubMed] [Google Scholar]
- 32.Yang, H., Zhong, X., Ballar, P., Luo, S., Shen, Y., Rubinsztein, D. C., Monteiro, M. J., and Fang, S. (2007) Exp. Cell Res. 313 538-550 [DOI] [PubMed] [Google Scholar]
- 33.Pye, V. E., Beuron, F., Keetch, C. A., McKeown, C., Robinson, C. V., Meyer, H. H., Zhang, X., and Freemont, P. S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 467-472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hocking, L. J., Lucas, G. J., Daroszewska, A., Mangion, J., Olavesen, M., Cundy, T., Nicholson, G. C., Ward, L., Bennett, S. T., Wuyts, W., Van Hul, W., and Ralston, S. H. (2002) Hum. Mol. Genet. 11 2735-2739 [DOI] [PubMed] [Google Scholar]
- 35.Pankiv, S., Clausen, T. H., Lamark, T., Brech, A., Bruun, J. A., Outzen, H., Overvatn, A., Bjorkoy, G., and Johansen, T. (2007) J. Biol. Chem. 282 24131-24145 [DOI] [PubMed] [Google Scholar]
- 36.Skibinski, G., Parkinson, N. J., Brown, J. M., Chakrabarti, L., Lloyd, S. L., Hummerich, H., Nielsen, J. E., Hodges, J. R., Spillantini, M. G., Thusgaard, T., Brandner, S., Brun, A., Rossor, M. N., Gade, A., Johannsen, P., Sorensen, S. A., Gydesen, S., Fisher, E. M., and Collinge, J. (2005) Nat. Genet. 37 806-808 [DOI] [PubMed] [Google Scholar]
- 37.Filimonenko, M., Stuffers, S., Raiborg, C., Yamamoto, A., Malerod, L., Fisher, E. M., Isaacs, A., Brech, A., Stenmark, H., and Simonsen, A. (2007) J. Cell Biol. 179 485-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J. A., Beigneux, A., Ahmad, S. T., Young, S. G., and Gao, F. B. (2007) Curr. Biol. 17 1561-1567 [DOI] [PubMed] [Google Scholar]
- 39.Nishino, I. (2003) Curr. Neurol. Neurosci. Rep. 3 64-69 [DOI] [PubMed] [Google Scholar]
- 40.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y., and Shimizu, N. (1998) Nature 392 605-608 [DOI] [PubMed] [Google Scholar]
- 41.Olzmann, J. A., Li, L., Chudaev, M. V., Chen, J., Perez, F. A., Palmiter, R. D., and Chin, L. S. (2007) J. Cell Biol. 178 1025-1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan, J. M., Wong, E. S., Kirkpatrick, D. S., Pletnikova, O., Ko, H. S., Tay, S. P., Ho, M. W., Troncoso, J., Gygi, S. P., Lee, M. K., Dawson, V. L., Dawson, T. M., and Lim, K. L. (2008) Hum. Mol. Genet. 17 431-439 [DOI] [PubMed] [Google Scholar]