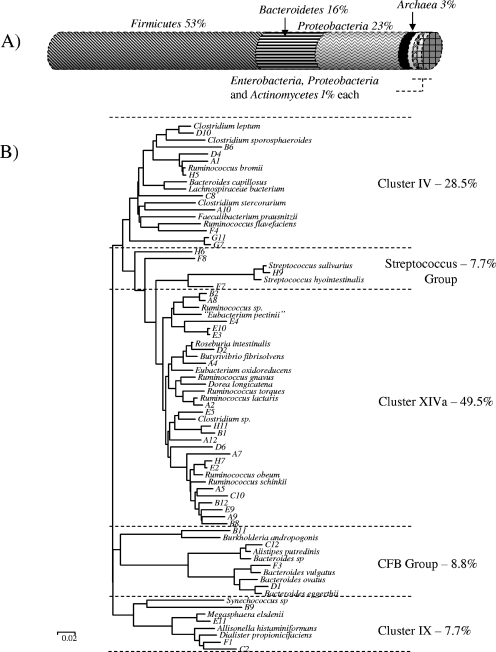
FIG. 3.
(A) Diagram depicting the results of sequencing each end of some of the Tcr fosmid clones isolated from the Ab1 metagenomic fosmid library by use of vector-specific primers. Sequences were analyzed using the program BlastX at the network service hosted by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Results were distributed within one of the bacterial phyla shown and expressed as percentages. For comparison, bacteria included in phylogroups IV, IX, and XIVa and the streptococcus group (below) all belong to the phylum Firmicutes. (B) Phylogenetic analysis of 96 cloned 16S rRNA sequences amplified using eubacterial universal primers from DNA extracted from the Ab1 fecal sample. The phylogenetic tree (rooted phylogram) was created using the ClustalX neighbor-joining method and edited using MEGA 3.1. The number of bootstrap trials was set to 1,000. The proportion of bacterial species falling into each of the five phylogroups detected is shown. CFB, Cytophaga-Flavobacterium-Bacteroidetes group, corresponds to Bacteroidetes in panel A.