Abstract
The human pathogenic poxvirus molluscum contagiosum virus (MCV) is the causative agent of benign neoplasm, with worldwide incidence, characterized by intraepidermal hyperplasia and hypertrophy of cells. Here, we present evidence that the MC007L protein of MCV targets retinoblastoma protein (pRb) via a conserved LxCxE motif, which is present in many viral oncoproteins. The deregulation of the pRb pathway plays a central role in tumor pathogenesis. The oncoproteins of small DNA viruses contain amino acid sequences that bind to and inactivate pRb. Isolated expression of these oncoproteins induces apoptosis, cell proliferation, and cellular transformation. The MC007L gene displays no homology to other genes within the poxvirus family. The protein anchors into the outer mitochondrial membrane via an N-terminal mitochondrial targeting sequence. Through the LxCxE motifs, MC007L induces a cytosolic sequestration of pRb at mitochondrial membranes, leading to the inactivation of the protein by mislocalization. MC007L precipitates the endogenous pRb/E2F-1 complex. Moreover, MC007L is able to cooperate to transform primary rat kidney cells. The interaction between MC007L and pRb provides a novel mechanism by which a virus can perturb the cell cycle.
Molluscum contagiosum virus (MCV), a member of the family Poxviridae, causes intraepidermal hyperplasia in humans, especially immunodeficient patients. MCV and variola (smallpox) virus are the only known poxviruses that are specific for the human host. In contrast to the acute generalized infection with high mortality caused by variola virus, MCV produces a chronic infection localized to epidermal tissue (10, 27). This is characterized by hyperplasia and hypertrophy of the virus-infected keratinocytes (7, 35, 37), and MCV-infected tissue shows an abnormal expression of proteins of the cytoskeleton and cell cycle (35, 37). In the absence of a functional immune system, MCV causes giant molluscum, with extensive disfiguring lesions (19). MCV does not complete the replication cycle in cell culture; however, some early and late genes were expressed (1, 2). As for all poxviruses, viral replication occurs in the cell cytoplasm (7).
Uncontrolled cellular proliferation as a consequence of cell cycle deregulation is the major cause of tumors. The retinoblastoma protein (pRb)/E2F complex plays a central role in the control of cell proliferation (23, 40). The functions of pRb relative to cell cycle progression are mediated by the protein's binding to and inactivation of the transcription factors of the E2F family (12, 13). The E2F factors coordinate the transcription of a large family of proteins essential for cell cycle progression and apoptosis (12, 29). Diverse viral oncoproteins, such as the human papillomavirus type 16 (HPV16) E7 protein, the simian virus 40 (SV40) large T (LT) antigen, and adenovirus E1a all bind the pocket domain of the pRb family of proteins, pRb, p107, and p130, mainly through an LxCxE motif present within their conserved region 2 (CR2) domains. The disruption of the pRb/E2F protein complex leads to cell proliferation, induction of apoptosis, and cellular transformation (6, 8, 15, 38). Several MCV unique open reading frames (ORFs) predicted to have a host-interactive function were experimentally characterized and found to protect cells from apoptosis (16, 22, 33, 34). Since determination of the primary structure of the MCV genome (30, 31), the functional activities of many important viral genes have been identified and characterized, including that for MC148R, with homology to members of the beta or CC family of chemokines (3, 17, 18, 32, 42). In addition, a number of MCV ORF proteins with no counterparts in other poxviruses revealed significant homology to cellular proteins. Like other viruses, MCV possesses proteins MC159 and MC066, known as immune defense molecules producing favorable conditions for viral replication (22, 32, 33, 34).
Starting from the hypothesis that MCV possesses a gene(s) which interferes with proteins regulating the cell cycle, viral ORF proteins were screened for the presence of a pRb-binding motif, LxCxE, which is found in a number of cell cycle regulatory proteins and in some viral oncoproteins (5, 6, 8). Our findings show that the MC007L gene of human pathogenic MCV encodes a mitochondrial outer membrane (MOM) protein that targets pRb and E2F-1. MC007L induces a mitochondrial sequestration that leads to the inactivation of pRb, and this binding capacity provides MC007L with the ability to cooperate with Ras to transform primary baby rat kidney (BRK) cells. These findings reflect a novel mechanism by which a virus can interfere with the cell cycle.
MATERIALS AND METHODS
Cell culture.
CV1, primary BRK cells, Saos-2, U2OS, and HeLa cells are maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg/ml penicillin, and 100 U/ml streptomycin (Invitrogen).
Transformation assay.
Kidneys were obtained from 9-day-old Wistar rats and transfected with the relevant plasmid combinations using calcium phosphate precipitation essentially as described previously (21). Cells were placed under G418 selection (200 μg/ml) and left for 2 to 3 weeks. After this time, the colonies obtained were counted and polyclonal pools were generated for subsequent analysis.
Expression vectors.
To construct a full-length MC007L plasmid, the MC007L ORF was amplified by PCR using the forward primer 007S, 5′GGAGAATTCATGCATGCGGAGCTGGCCGAAGTC3′, cut with EcoRI, and reverse primer 007AS-FLAG, 5′GCCTCTAGACTACTTGTCGTCGTCGTCCTTATAGTCCGCCTCCGCGCGTGCAAG3′, containing the FLAG M2 oligonucleotide sequence, cut with XbaI, selected from the genomic MCV sequence described in reference 31 (GenBank accession number U60315; genome location 12614 to 13264 bp). Genomic viral DNA isolated from MCV served as a template. The amplified sequence was cloned by EcoRI-XbaI digestion into the pCR3 vector (Invitrogen). To generate pCR3-MC007L-143/180-FLAG, site-directed mutagenesis of the two central cysteines within the pRb-binding LxCxE motifs of MC007L was performed by PCR. The forward primers described below were used (codons with mutated nucleotides are indicated). The mutant with a cysteine-to-glycine mutation at amino acid position 143 was constructed using the forward primer C143G-S, 5′TGCGTGGACGTGGACCTCTACGGTCACGAGAACTTGCGCTACGAG3′ (+), and cloned into an EcoRI- and XbaI-digested pCR3 vector. The mutant with a cysteine-to-glycine mutation at amino acid position 180 was generated with the forward primer (C180G-S), 5′ACACTTCTGGATCTGCTCGCGGGCTCCGAGGACGCGAGCGGGTTTTCG3′(+), and an MC007L-C143G mutant construct served as a template. The double mutant construct was cloned via EcoRI-XbaI digestion into pCR3. The construct pCR3-MC007L-M54L-FLAG was generated with the mutagenesis primer using the following primers: forward (M54L-S), 5′CGGCACTGCGTACACTTGGCGGCGCGC3′; and reverse (M54L-AS), 5′CGCCGCCGCCAAGTGTAGGCAGTGCCG3′. The construct pCR3-MC007L-Δ(1-53)-FLAG was generated using the forward primer 007-53S, 5′GGAGAATTCATGGCCGCGCGCTACTTGTCG 3′, and the reverse primer 007AS-FLAG, containing the FLAG sequence as described above. The amplified DNA was cloned via EcoRI/XbaI digestion into pCR3.
For construction of MC007L-RFP and MC007L-143/180RFP, the monomeric red fluorescent protein was amplified by PCR from the plasmid pRSETb (4) using the forward primer RFP-S, 5′GACTCTAGAGCTAGCATCACTGGTGGACAG3′, and the reverse primer RFP-AS, 5′CGGTCTAGATTAGCCGCCGGTGGAGTGGCGGCC3′. The red fluorescent protein fragment was cloned via XbaI in frame with MC007L or MC007L-143/180 into pCR3 plasmid. The constructs were confirmed by sequencing.
The plasmids Myc-pRb, Myc-pRb-SP and Myc-pRb 756/757 were kindly provided by S. Mittnacht; E2F-1 was provided by J. R. Nevins.
Isolation of MCV and infection procedure.
Cells were infected with MCV prepared from patient lesions. Pooled lesions were homogenized in ice-cold phosphate-buffered saline (PBS) and centrifuged at 6,000 × g to remove cell debris. Virus-containing supernatant was analyzed by negative-contrast electron microscopy to determine the virus morphology (data not shown). CV1 cells were incubated with MCV for 3 hours. Following infection, fresh medium was added. The immunofluorescence was performed after an additional incubation for 24 h.
Transfection experiments.
Cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100 μg/ml penicillin, and 100 U/ml streptomycin (Invitrogen). Transient cotransfections were performed with FuGENE6 (Roche) according to the manufacturer's instructions using the MC007L plasmids or mutant constructs. The immunofluorescence was performed as described previously (43).
Immunofluorescence and confocal microscopy.
Immunofluorescence studies were performed as described in reference 43. In brief, cells were fixed with 100% methanol at −20°C or 3% paraformaldehyde, washed with PBS, and blocked with 0.5% gelatin. Cells were stained with the following antibodies: anti-pRb monoclonal antibody (MAb) clone G3-245, cytochrome c (Cyt c), and clone 6H2.B4 (BD Biosciences). To detect MC007L, a polyclonal rabbit serum raised against a synthetic peptide, CSEDASGFSPPEDSFS, was generated and affinity purified (Eurogentec). Additionally, MC007L was detected using an anti-M2 FLAG antibody (Sigma). The optimal detection and separation of individual fluorescence was performed by sequential scan mode on a Leica TCS SP2-AOBS confocal laser scanning microscope (Bensheim, Germany). Apoptosis was detected using the in situ cell death detection kit, TMRred (Roche). In brief, CV1 or U2OS cells were transiently transfected with either the empty vector or the construct expressing MC007L-FLAG. Cells were washed in PBS and fixed with 4% paraformaldehyde for 1 h. Cells were then permeabilized with 0.1% Triton X-100 for 5 min. Coverslips were rinsed in PBS and labeled with 50 μl of terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction mixture per sample. To generate the positive control, transfected cells were incubated with DNase I prior to TUNEL. For MC007L detection, cells were immunostained with a MAb specific for FLAG and subsequently labeled with fluorescein isothiocyanate (FITC) conjugate. Cell nuclei were analyzed with confocal microscopy.
To determine the pRb distribution, 200 of the MC007L transfected cells were inspected and the intracellular localization of pRb was determined. Two independent transfection experiments were documented.
Isolation of mitochondria.
HeLa cells were seeded on a 15-cm plate at 80% confluence 1 hour before classical calcium phosphate transfection. Cells were harvested 36 h later by trypsinization, and the cell pellet was washed with MB buffer (210 mM mannitol, 70 mM sucrose, 1 mM EDTA, and 10 mM HEPES [pH 7.5]). Cells were broken with 30 strokes using a Dounce-Potter homogenizer and centrifuged at 500 × g for 5 minutes at 4°C. This procedure was repeated until almost all of the cells were broken. The supernatant obtained was spun at 2,000 × g to pellet the nucleus and then at 10,000 × g for 5 minutes to pellet the mitochondria. Mitochondria were washed once and resuspended in MB buffer. Protein concentration was estimated by the Bradford method (Bio-Rad).
Proteinase K assay.
A total of 100 μg of the mitochondria was incubated in 200 μl of MB buffer with proteinase K (Roche) at the indicated concentration for 30 min at 37°C. The digestion was stopped after the addition of 1 mM phenylmethylsulfonyl fluoride. The digested fraction was subsequently centrifuged at 10,000 × g for 5 min at 4°C. The pellet was further resuspended in 50 μl of 1× sodium dodecyl sulfate (SDS) loading buffer and separated by SDS-polyacrylamide gel electrophoresis (PAGE) for analysis by immunoblotting.
Sodium carbonate treatment.
The mitochondrial pellet was suspended in 0.1 M Na2CO3, pH 12, to a final concentration of 0.5 mg/ml and incubated on ice for 20 min. The sample was further centrifuged at 100,000 × g for 30 min. The pellet and the supernatant, classically precipitated with TCA, were resuspended in 50 μl of 1× SDS loading buffer and then separated by SDS-PAGE for analysis by immunoblotting.
Immunoprecipitation and immunoblotting.
Immunoprecipitation was performed as described previously with the following modifications: cells were lysed 48 h posttransfection with the precipitation buffer (1% of Triton X-100, 150 mM to 200 mM NaCl, 50 mM NaF, 1 mM sodium orthovanadate, 20 mM Tris [pH 7.5], 1 mM EDTA, and 0.2 mM phenylmethylsulfonyl fluoride, with a complete set of protease inhibitors [Sigma]) (43). Lysates were precleared with protein A Sepharose beads (Amersham). Whole lysates were precipitated with a polyclonal anti-FLAG M2 antibody for 4 hours at 4°C. Immune complexes were washed twice with a precipitation buffer, followed by PBS. One-tenth of the total lysate and the immunoprecipitated samples was subjected to SDS-PAGE. The immunoblot analysis was performed using MAb specific for pRb (clone G3-245; BD Bioscience), E2F-1 (clone KH95 or C-20; Santa Cruz Biotechnology), Myc (clone 9E10; Santa Cruz Biotechnology), and β-actin (Sigma). The following mitochondrial compartment markers were used: for the inner mitochondrial membrane, an anti-SLP2 antibody was produced by immunizing rabbits with a synthetic peptide (J. C. Martinou); for the mitochondrial matrix, the monoclonal anti-HSP70 (Affinity BioReagents) antibody was used; and for detection of MOM, the polyclonal anti-hFis1 antibody (Apotech) was applied.
RESULTS
MC007L is localized to the mitochondria.
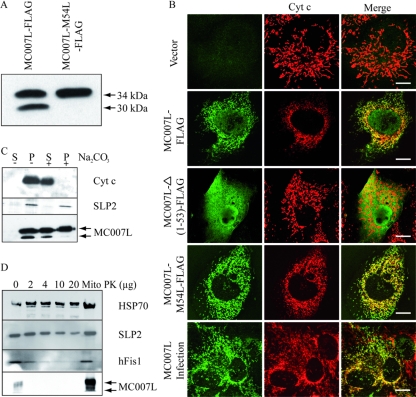
MCV possesses genes with no homology to those of other poxviruses, suggesting unique functions in manipulating host cell physiology. To identify new MCV functions, some of these ORF proteins were screened for the presence of the amino acid sequence motif LxCxE. We identified the MC007L ORF protein, which is 217 amino acids in length and has two LxCxE motifs, the first being embedded in a conserved sequence found within the HPV16 E7, SV40 LT, and adenovirus E1a oncoproteins (Fig. 1A and B). Interestingly, while no mitochondrial localization was predictable (http://psort.ims.u-tokyo.ac.jp/), an N-terminal hydrophobic amino acid sequence of 19 residues was predicted to be a transmembrane domain (TMD), suggesting probable membrane localization (Fig. 1A and C). Furthermore, the TMD is flanked by positively charged amino acids as described for N-terminally anchored proteins of the MOM, for example, Tom20 (translocase of the outer mitochondrial membrane), which is important for sorting and anchoring proteins into the MOM (Fig. 1C) (20, 28). Transient expression of MC007L-FLAG revealed two protein products of 30 and 34 kDa instead of one protein product. The larger MC007L-FLAG differs from the calculated molecular mass of 24 kDa. Replacing the internal methionine by a leucine at position 54 resulted in the expression of a single 34-kDa protein. Thus, the faster-migrating protein results from internal initiation of translation (Fig. 2A). Therefore, we generated the faster-migrating MC007L which lacks the N-terminal amino acid residues 1 to 53.
FIG. 1.
Amino acid sequence of the MC007L gene product. (A) Amino acid sequence of MC007L ORF protein. MC007L contains a short amino-terminal hydrophobic sequence (underlined) flanked by positively charged amino acids (bold). The CR2 domain and the LxCxE pRb binding motifs are shown in gray and bold. The peptide sequence used to generate serum against MC007L is shown in italics. (B) Sequence homology between CR2 of MC007L and adenovirus (Ad5) E1a, E7 of HPV16, and SV40 LT antigen. The amino acid position of the CR is indicated as follows: MC007L (134 to 164), E1a (115 to 143), E7 (15 to 42), and LT (96 to 123). A gap of 1 amino acid was introduced in the LT CR2 sequence (6). (C) The amino-terminal tail of MC007L contains a hydrophobic sequence flanked by a positively charged amino acid similar to that of the MOM resident protein Tom20. H.s., Homo sapiens.
FIG. 2.
The MCV protein MC007L localizes to mitochondria. (A) Transient expression of MC007L and MC007L-M54L-FLAG constructs. The FLAG-tagged constructs were expressed transiently in CV1 cells. After 24 h, cells were lysed and subjected to immunoblot analysis. MC007L was detected using a FLAG-specific antibody. (B) MC007L colocalizes with the mitochondrial marker Cyt c. CV1 cells were transfected with either an empty vector (first row), MC007L-FLAG (second row), MC007L-Δ(1-53)-FLAG (third row), or MC007L-M54L-FLAG (fourth row). At day 1 posttransfection, the FLAG-tagged MC007L proteins were detected using a rabbit anti-FLAG or a monoclonal anti-Cyt c antibody. The primary antibodies were labeled using FITC or Cy3. The immunofluorescence was analyzed by confocal microscopy. Bar, 15 μm. MC007L localizes to the mitochondria in MCV-infected cells. CV1 cells were infected with MCV and subjected to immunofluorescence 24 h later. To detect the viral protein, a peptide serum raised against MC007L was used. The mitochondria were visualized using an anti-Cyt c antibody. Bar, 20 μm. (C) MC007L-FLAG is inserted into the mitochondrial membrane. HeLa cells were transfected with the MC007L-FLAG construct and fractionated into mitochondrial membranes and cytosolic fraction. The mitochondrial pellet from MC007L-FLAG transfected HeLa cells were extracted for 20 min with Na2CO3. The mitochondrial pellet and supernatant were subjected to immunoblot analysis. MC007L was detected using a rabbit antiserum. SLP2 is a marker for the inner mitochondrial membrane, and HSP70 resides in the mitochondrial matrix. (D) MC007L is an MOM protein. Purified mitochondria (Mito) were incubated with increasing concentrations of proteinase K (PK) (shown in μg). After proteinase K digestion, fractions were analyzed by immunoblot analysis. The following mitochondrial markers were used: HSP70 for the mitochondrial matrix, SLP2 for the mitochondrial inner membrane, and hFis1 for the MOM.
To investigate its cellular localization, we expressed C-terminally FLAG-tagged MC007L in CV1 cells. MC007L-FLAG colocalized with the mitochondrial marker Cyt c in transient transfection assays (Fig. 2B, second row), but not with the endoplasmic reticulum marker calnexin (data not shown). Nuclear staining was also observed, suggesting that the faster-migrating MC007L-FLAG localized to the nucleus. As a negative control, the empty vector was transfected into CV1 cells (Fig. 2B, first row). The faster-migrating MC007L-Δ(1-53)-FLAG construct revealed a diffuse, cytosolic, and nuclear pattern of distribution (Fig. 2B, third row). In order to clarify the MC007L localization, the MC007L-M54L-FLAG mutant construct was transiently transfected into CV1 cells and analyzed with confocal microscopy using Cyt c as a mitochondrial marker. The mutated long form (with a mutated methionine at position 54) (Fig. 1A) localized at the mitochondria was shown by colocalization with Cyt c (Fig. 2B, fourth row). Thus, the residues 1 to 53 of MC007L are involved in the localization of MC007L to the mitochondria.
Although MCV does not undergo a complete viral life cycle in cell culture associated with virus morphogenesis, some early and late viral genes are expressed (1, 2). To verify that MC007L is expressed by the virus and exhibits a cellular distribution similar to that observed in transfected cells, CV1 cells were infected with MCV and analyzed 24 h later for the pattern of MC007L localization in combination with Cyt c. The immunofluorescence analysis revealed a mitochondrial distribution of the MC007L protein in MCV-infected cells. Thus, MC007L colocalized with Cyt c during infection (Fig. 2B, fifth row).
To detect the viral protein, a polyclonal rabbit serum raised against a synthetic peptide (see Materials and Methods) was generated. To test the specificity of the antiserum, CV1 cells were transfected with the MC007L-FLAG construct and subjected to immunofluorescence using the MAb specific for FLAG and a rabbit serum against the MCV protein. Confocal microscopy analysis revealed a merge between the two stainings, suggesting the specificity of the peptide serum (data not shown). Importantly, MC007L expressed in isolation does not induce a release of proapoptotic mitochondrion-associated Cyt c (Fig. 2B), nor does it induce positive TUNEL staining in CV1 or U2OS cells (data not shown).
To further confirm the mitochondrial membrane association of MC007L, HeLa cells were transiently transfected and subjected to subcellular fractionation after 24 h. Alkaline treatment of purified mitochondrial fraction showed a partial extraction of the MC007L-FLAG protein which cofractionates with the mitochondrial membrane marker stomathin-like protein 2 (Fig. 2C). However, the faster-migrating MC007L cofractionated with the mitochondria is completely washed out by the addition of sodium carbonate. This complete depletion suggests that the shorter protein does not insert into the mitochondrial membrane (Fig. 2C). To determine whether MC007L inserts into the MOM, the purified mitochondrial fraction was digested with increased amounts of proteinase K. The result shows a concomitant degradation of MC007L and hFis1, a known component of the MOM (Fig. 2D) (25). Altogether, these results demonstrate that the MC007L gene product is N-terminally anchored in the MOM.
MC007L targets pRb and E2F-1.
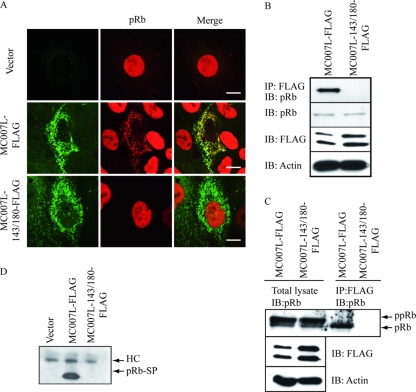
To test whether the LxCxE motif was functional and conferred pRb targeting to the mitochondria, CV1 cells were transiently transfected either with MC007L-FLAG or a mutant lacking the functional LxCxE motifs. Since both motifs are potentially capable of binding to pRb, we generated a double mutant, MC007L-143/180-FLAG, in which the central cysteine residue within the LxCxE motif was replaced by glycine. The double cysteine-to-glycine (C143G and C180G) mutant of MC007L-FLAG should fail to bind pRb but still localize to the MOM. MC007L-FLAG and the mutant revealed similar expression patterns in transiently transfected CV1 cells (Fig. 3A). CV1 cells were transiently transfected with either MC007L-FLAG or an MC007L-143/180-FLAG construct with functional mutations of the two LxCxE motifs. The endogenous pRb colocalized with MC007L-FLAG to mitochondria (Fig. 3A, second row) in up to 20% of transfected cells, whereas the mutant MC007L-143/180-FLAG failed to recruit it (Fig. 3A, third row). Importantly, cells with retained mitochondrial pRb were negative for TUNEL staining (data not shown) and revealed no Cyt c release. The small pocket domain of pRb-SP (379 to 792) (data not shown) and the pRb-ΔNLS (data not shown) are both retained at the mitochondria in all MC007L-FLAG-expressing cells, indicating that the LxCxE motifs protrude into the cytosol and are accessible to target pRb.
FIG. 3.
The poxvirus protein MC007L interacts with the endogenous pRb. (A) MC007L recruits the endogenous pRb to mitochondria in CV1 cells. CV1 cells were transiently transfected with either MC007L-FLAG (second row) or MC007L-143/180-FLAG (third row) plasmid for 48 h. After fixation, cells were subjected to immunofluorescence. MC007L was detected using an anti-FLAG antibody and pRb was detected with the MAb G3-245. As a negative control, an empty vector was used (first row). The immunofluorescence was analyzed by confocal microscopy. For double staining, a combination of two secondary antibodies was applied, an FITC conjugate and a Cy3 conjugate. Bar, 15 μm. (B) MC007L coimmunoprecipitates endogenous pRb. CV1 cells were transfected with MC007L-FLAG or MC007L-143/180-FLAG, and after 48 h the cell lysate was subjected to coimmunoprecipitation using a rabbit anti-FLAG antibody. Detection of precipitates was performed by immunoblot analysis, and the antibodies used are indicated below. One-tenth of the total lysate was used as a loading control. (C) MC007L binds the endogenous hypophosphorylated pRb. Cell lysate from transfected CV1 cells was precipitated with an anti-FLAG antibody and subjected to immunoblot staining using a monoclonal anti-pRb antibody G3-245. One-tenth of the loading control is shown in the left panel. (D) MC007L coimmunoprecipitates the small pocket of pRb. CV1 cells were transfected with MC007L-FLAG, MC007L-143/180-FLAG, or the empty vector in combination with the pRb-SP Myc-tagged construct, and after 24 h the cell lysate was subjected to coimmunoprecipitation using a rabbit anti-FLAG antibody. Detection of precipitates was performed by immunoblotting, and the small pocket domain was detected using a specific antibody against Myc (clone 9E10). HC, heavy chain.
The interaction of MC007L-FLAG with endogenous pRb was confirmed by coimmunoprecipitation. MC007L-FLAG or MC007L-143/180-FLAG was transfected into CV1 cells and subjected to immunoprecipitation 48 h later. The lysate was precipitated with a polyclonal antibody to FLAG, and pRb was detected using G3-245 antibody. Whereas the wild-type MC007L binds the endogenous pRb, the MC007L LxCxE motif mutant failed to bind (Fig. 3B). For reciprocal coimmunoprecipitation, the monoclonal (G3-245) (data not shown) or a polyclonal (C-15) serum against pRb was used. To detect MC007L, an anti-M2 FLAG antibody was applied. The interaction of the two proteins could not be confirmed by reciprocal coimmunoprecipitation (data not shown).
To test whether the viral protein coimmunoprecipitates the hypophosphorylated pRb, MC007L and the LxCxE motif mutant were transfected into CV1 cells. Cell lysates were precipitated 48 h after transfection, using an anti-M2 FLAG antibody, and pRb was detected with the pRb-specific monoclonal G3-245 antibody. Low-percentage polyacrylamide gel allows for distinguishing between the hypo- and hyperphosphorylated pRb forms. MC007L precipitated the hypophosphorylated pRb, whereas the LxCxE motif mutant failed to coimmunoprecipitate (Fig. 3C). This result is in agreement with other viral oncoproteins. To further confirm the interaction between pRb and MC007L, a construct expressing the small pocket domain of pRb tagged with Myc was cotransfected with the MC007L plasmid. The wild-type MC007L coprecipitated the pRb-SP domain, whereas the viral protein mutant with mutations of the LxCxE motifs failed to do so (Fig. 3D). As mentioned above, this result could be confirmed by colocalization studies (data not shown). Importantly, the pRb-SP was retained at mitochondria in all MC007L-expressing cells.
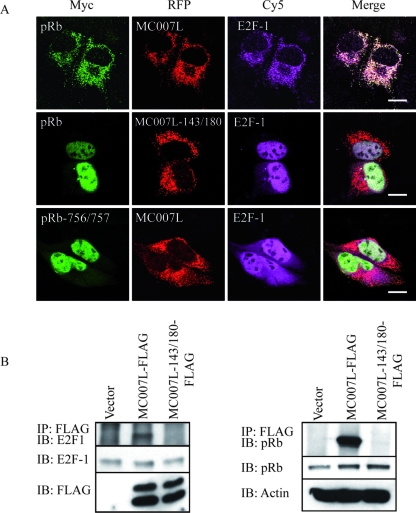
Since E2F-1 is one of the main targets of pRb, we analyzed the intracellular distribution of ectopically expressed E2F-1 together with that of pRb in cells expressing the viral protein. Saos-2 cells were transiently transfected with either MC007L-RFP or MC007L-143/180-RFP, together with the Myc-tagged full-length pRb and E2F-1 plasmid, for 24 h. Cells were subjected to immunofluorescence with a MAb against pRb and rabbit antibody against E2F-1. Cells expressing MC007L-RFP that induced pRb relocation also retained E2F-1 at the mitochondria (Fig. 4A, first row). The MC007L deficient in pRb binding with mutated LxCxE motifs failed to retain pRb and also failed to retain E2F-1 at the mitochondria (Fig. 4A, second row) showed by nuclear localization. Moreover, the pRb mutant 756/757 construct, full-length but LxCxE binding defective, also failed to target pRb (Fig. 4A, third row). Thus, the targeting of pRb and E2F-1 is LxCxE motif dependent in transient Saos-2 expression systems.
FIG. 4.
MC007L interacts with the pRb/E2F-1 complex. (A) MC007L targets pRb and E2F-1 at the mitochondria in transient transfection assays. Saos-2 cells were transiently transfected with MC007L-RFP (first row) or with the MC007L mutant with mutated LxCxE motifs (second row) together with Myc-tagged pRb and E2F-1-expressing plasmids. After 24 h posttransfection, cells were fixed and stained with a MAb specific for Myc and a rabbit anti-E2F-1 antibody. In the third row, Saos-2 cells transfected with MC007L-RFP together with a Myc-tagged pRb 756/757 and E2F-1 plasmids were subjected to immunofluorescence 24 h posttransfection. Cells were fixed and stained with a MAb specific for Myc and a rabbit anti-E2F-1 antibody. Two secondary antibodies were applied with FITC or Cy5, respectively. Bar, 15 μm. (B) MC007L precipitates the endogenous pRb/E2F-1 complex. MC007L coimmunoprecipitates the endogenous pRb/E2F-1 complex. CV1 cells transfected with empty vector, MC007L-FLAG, or MC007L-143/180-FLAG were subjected to coimmunoprecipitation using an anti-FLAG-specific antibody. Precipitates were analyzed by immunoblot analysis using the antibodies indicated below. MC007L was detected using an anti-FLAG antibody and E2F-1 was detected with the MAb KH95. To detect endogenous pRb, the blot was reprobed with the anti-pRb-specific antibody G3-245 and β-actin-specific MAb, respectively. One-tenth of the loading control was used.
We next determined whether the endogenous E2F-1 forms a complex in cells expressing MC007L, concomitant with endogenous pRb. CV1 cells were transiently transfected with either MC007L-FLAG or MC007L-143/180-FLAG for 24 h. Lysates were precipitated with an antibody against FLAG and separated by SDS-PAGE. The immunoblot was probed with an anti-E2F-1 antibody and reprobed with anti-pRb antibody. As can be seen, the MC007L-FLAG binding to endogenous E2F-1 is LxCxE dependent (Fig. 4B), since efficient coimmunoprecipitation is obtained with wild-type MC007L; the MC007L mutant, while failing to bind the pRb, is unable to coprecipitate E2F-1.
MC007L cooperates with EJ-ras to transform primary BRK cells.
The viral oncoproteins adenovirus E1A, SV40 LT antigen, and HPV16 E7 are able to induce cell transformation which, in cooperation with an activated EJ-ras oncoprotein, is dependent upon their pRb-binding capacities. To investigate whether MC007L possesses a cell transformation capacity, primary BRK cells were transfected with MC007L-FLAG or MC007L-143/180-FLAG mutant plasmids, together with EJ-ras and posttransfection maintained under antibiotic selection for a period of 2 to 3 weeks. Transformed colonies were obtained only from the MC007L protein. The colonies were counted (Fig. 5), and polyclonal pools generated were tested for the presence of MC007L by immunoblot analysis. All cells expressed the viral protein (data not shown). Taken together, MC007L-FLAG in cooperation with Ras was able to transform rodent cells, in contrast to the MC007L mutant in LxCxE motifs, which failed to do so (Fig. 5). The transformation assay suggests a modest ability of MC007L to contribute to the transformation of primary BRK cells in the presence of Ras.
FIG. 5.
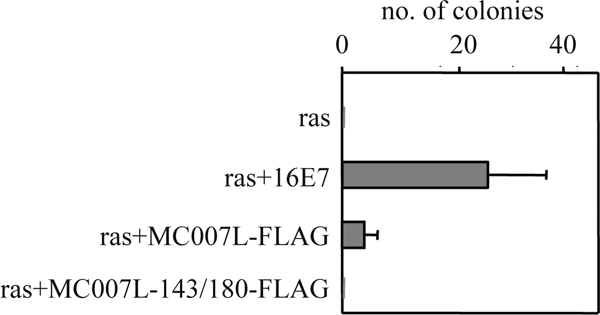
MC007L-FLAG cooperates with EJ-ras to transform primary BRK cells. BRK cells were transfected with MC007L-FLAG or MC007L-143/180-FLAG constructs and incubated for 3 weeks in the presence of G418. Transformed colonies were counted and pooled for further analysis.
DISCUSSION
In this report, we show that the human pathogenic poxvirus MCV possesses an MC007L ORF protein, which contains two pRb-binding LxCxE motifs; one lies within a CR2-like domain, and the second is localized C terminally. An N-terminal mitochondrion-targeting sequence targets MC007L to the MOM, a finding that contradicts the computational prediction, which failed to indicate its localization. The viral protein induces a sequestration of endogenous pRb via the two LxCxE motifs at the mitochondria. Because all viral oncoproteins contain only one LxCxE motif, we assume that one of the motifs is sufficient to target pRb. Moreover, the endogenous E2F-1 sequestration depends on the LxCxE motif within the pRb protein. In addition, MC007L is modestly able to cooperate with Ras to transform primary rat cells (Fig. 5). In contrast to classical viral oncoproteins, MC007L does not appear to induce apoptosis as determined by TUNEL assays in CV1 and U2OS cells. However, the mitochondrial localization of MC007L strongly suggests an additional function in regulation of apoptosis, without or with additional MCV proteins, e.g., MC159 or MC160 (24, 33, 34).
To date, all viral proteins that have a mitochondrial localization exhibit functions that can affect apoptosis. The mitochondrially associated poxviral proteins, M11L from myxoma virus and F1L from vaccinia virus, both of which have anti-apoptotic functions, insert into the mitochondria via their C termini (11, 36, 39). These proteins possess at their C termini a hydrophobic TMD flanked by positively charged residues (36). Our fractionation and colocalization studies on MC007L with a variety of mitochondrial markers suggest that MC007L inserts into the MOM through its N terminus, similar to Tom20 or hFis1 (Fig. 1C and 2D). The proteinase K sensitivity is similar to that of hFis1, a protein known to localize at MOM (Fig. 2D) (25). Alkaline extraction of the mitochondria resulted in a complete loss of the MC007L-Δ(1-53)-FLAG, thereby confirming that the shorter protein does not insert into the MOM. While the SLP2 protein, a marker for the inner mitochondrial membrane, is resistant against carbonate extraction, MC007L is partially extracted from the membrane. This could be due to overexpression of the protein and the fact that some of it is not completely inserted, or MC007L may require for membrane insertion the Tom complex that is saturated due to overexpression of the protein (Fig. 2C). The cofractionation of the shorter MC007L-Δ(1-53)-FLAG protein with the mitochondrial fraction (Fig. 2C) may also suggest that MC007L can interact with itself or with a protein located at the MOM. A reciprocal coimmunoprecipitation with MC007L-FLAG or MC007L-hemagglutinin confirmed that MC007L can at least dimerize (data not shown). The findings that a moderate hydrophobicity of the TMD and the presence of positively charged residues in the C-terminal flanking region target Tom20 to the MOM (20, 28). We propose a similar mitochondrial targeting mechanism for MC007L. The role of the positively charged residues in the flanking region of MC007L in the mitochondrial targeting remains to be characterized in detail. Taken together, MC007L anchors with the N terminus in the MOM and the two pRb-binding motifs protrude into the cytosol.
A unique feature of the MC007L is its mitochondrial localization combined with the presence of pRb-binding motifs, motifs which are reserved primarily for nuclear proteins involved in cell cycle regulation (5, 9, 26, 41). The CR2 domain (Fig. 1A) of MC007L reveals similarity to the high-risk HPV E7 sequence, suggesting a high affinity in pRb binding. Like the viral oncoproteins, MC007L possesses an LxCxE motif within a CR2-like domain (Fig. 1A and B) (8, 26), but unlike the viral oncoproteins, it has an additional LxCxE motif located C terminally (Fig. 1A). While the CR2 domain of MC007L shows significant homology to E7 of HPV16, a CR1 motif is missing. The E7 CR1 does not bind pRb but inhibits the access of E2F-1 to pRb (5, 14) and therefore precludes the pRb/E2F complex formation, thereby highlighting significant differences in how the two viral oncoproteins affect pRb function. In contrast, MC007L is able to bind to the pRb/E2F complex. Like the other viral oncoproteins, MC007L binds to the hypophosphorylated form of pRb (Fig. 3C). MC007L-FLAG is able to coimmunoprecipitate the small pocket domain of pRb in an LxCxE-dependent manner (Fig. 3D). Therefore, we considered this precipitation to be as valid as that achieved by glutathione S-transferase pull down assays.
Since MC007L is mitochondrially localized and pRb is a nuclear phosphoprotein, it would seem likely that the MC007L-pRb-E2F interaction occurs either during mitosis or immediately after synthesis of pRb, before it has translocated to the nucleus. The minority of MC007L-expressing cells exhibit a mitochondrial targeting of pRb, whereas the majority of cells show a nuclear localization of pRb. The fact that the small pocket protein domain of pRb (Fig. 3D; data not shown) and the pRb-ΔNLS mutant (data not shown) are retained to the mitochondria in all of the cells expressing MC007L suggests that the binding of MC007L to pRb can be restricted by importins or chaperones involved in the nuclear transport of pRb and that, under normal circumstances, only a portion of pRb is available for MC007L targeting. In addition, the influence of the cell cycle may play a role in targeting the pRb/E2F complex by MC007L. Nonetheless, the mitochondrially trapped pRb will be compromised in its function by this mislocalization in transient expression assays. Moreover, these cells were not found positive for TUNEL staining, and a Cyt c release was not detected. It is possible that cells with a mitochondrially targeted pRb/E2F-1 complex are also impaired in apoptotic function, since E2F-1 is a major regulator of apoptosis (29) and is inefficiently released from the trimeric complex MC007L-pRb-E2F, depending on the functional LxCxE motif (Fig. 4).
MC007L lacks the CR1 domain found in high- and low-risk E7 and is known to be involved in cellular transformation using activated Ras and BRK cells. The absence of the CR1 domain in MC007L might explain the decreased efficiency in cellular transformation. Moreover, the inefficient release of E2F-1 from the complex might also explain the low numbers of transformed BRK colonies obtained (Fig. 5). An important feature of the viral oncoprotein HPV16 E7 is the ability to induce tumors in vivo. HPV induces tumors, in contrast to MCV, which causes benign lesions of the skin even in immunocompromised individuals (19). However, there is clear proliferation of BRK cells (until passage 30).
The function of MC007L in the MCV context is precluded by the fact that MCV does not replicate in cell culture and does not exit an animal model. Nevertheless, MC007L represents a novel viral tool for studying the interference of viral proteins with the pRb/E2F complex and mitochondrial function.
Acknowledgments
We thank S. Mittnacht, J. Nevins, and R. Tsien for providing plasmids. We thank G. Giese for support in confocal laser scanning microscopy.
This work was supported in part by the Deutsche Forschungsgemeinschaft DA 142/10-1-5.
Footnotes
Published ahead of print on 13 August 2008.
REFERENCES
- 1.Bugert, J. J., C. Lohmüller, and G. Darai. 1999. Characterization of early gene transcripts of molluscum contagiosum virus. Virology 257119-129. [DOI] [PubMed] [Google Scholar]
- 2.Bugert, J. J., N. Melquiot, and R. Kehm. 2001. Molluscum contagiosum virus expresses late genes in primary human fibroblasts but does not produce infectious progeny. Virus Genes 2227-33. [DOI] [PubMed] [Google Scholar]
- 3.Bugert, J. J., C. Lohmüller, I. Damon, B. Moss, and G. Darai. 1998. Chemokine homolog of molluscum contagiosum virus: sequence conservation and expression. Virology 24251-59. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 997877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, H. M., L. Smith, and N. B. La Thangue. 2001. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 206152-6163. [DOI] [PubMed] [Google Scholar]
- 6.Chellappan, S., V. B. Kraus, B. Kroger, K. Münger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 894549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damon, I. K. 2007. Poxviruses, p. 2963-2965. In B. Knipe and D. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 8.Dyson, N., P. Guida, K. Münger, and E. Harlow. 1992. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 666893-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egan, C., T. N. Jelsma, J. A. Howe, S. T. Bayley, B. Ferguson, and P. E. Branton. 1988. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol. Cell. Biol. 83955-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein, W. L., and K. Fukuyama. 1973. Maturation of molluscum contagiosum virus (MCV) in vivo: quantitative electron microscopic autoradiography. J. Investig. Dermatol. 6073-79. [DOI] [PubMed] [Google Scholar]
- 11.Everett, H., M. Barry, S. F. Lee, X. Sun, K. Graham, J. Stone, R. C. Bleackley, and G. McFadden. 2000. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J. Exp. Med. 1911487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fattaey, A. R., E. Harlow, and K. Helin. 1993. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol. Cell. Biol. 137267-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heck, D. V., C. L. Yee, P. M. Howley, and K. Münger. 1992. Efficiency of binding the retinoblastomaprotein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc. Natl. Acad. Sci. USA 894442-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helin, K., E. Harlow, and A. Fattaey. 1993. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 136501-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helt, A. M., and D. A. Galloway. 2003. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis 24159-169. [DOI] [PubMed] [Google Scholar]
- 16.Hu, S., C. Vincenz, M. Buller, and V. M. Dixit. 1997. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J. Biol. Chem. 2729621-9624. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, Y., B. Wang, and F. D. Bushman. 1998. Molluscum contagiosum virus topoisomerase: purification, activities, and response to inhibitors. J. Virol. 723401-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa-Mochizuki, I., M. Kitaura, M. Baba, T. Nakayama, D. Izawa, T. Imai, H. Yamada, K. Hieshima, R. Suzuki, H. Nomiyama, and O. Yoshie. 1999. Molecular cloning of a novel CC chemokine, interleukin-11 receptor alpha-locus chemokine (ILC), which is located on chromosome 9p13 and a potential homologue of a CC chemokine encoded by molluscum contagiosum virus. FEBS Lett. 460544-548. [DOI] [PubMed] [Google Scholar]
- 19.Izu, R., D. Manzano, J. Gardeazabal, and J. L. Diaz-Perez. 1994. Giant molluscum contagiosum presenting as a tumor in an HIV-infected patient. Int. J. Dermatol. 33266-267. [DOI] [PubMed] [Google Scholar]
- 20.Kanaji, S., J. Iwahashi, Y. Kida, M. Sakaguchi, and K. Mihara. 2000. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matlashewski, G., J. Schneider, L. Banks, N. Jones, A. Murray, and L. Crawford. 1987. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. EMBO J. 61741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murao, L. E., and J. L. Shisler. 2005. The MCV MC159 protein inhibits late, but not early, events of TNF-α-induced NF-κB activation. Virology 340255-264. [DOI] [PubMed] [Google Scholar]
- 23.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10699-703. [DOI] [PubMed] [Google Scholar]
- 24.Nichols, D. B., and J. L. Shisler. 2006. The MC160 protein expressed by the dermatotropic poxvirus molluscum contagiosum virus prevents tumor necrosis factor alpha-induced NF-κB activation via inhibition of I kappa kinase complex formation. J. Virol. 80578-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parone, P. A., D. I. James, S. Da Cruz, Y. Mattenberger, O. Donzé, F. Barja, and J.-C. Martinou. 2006. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol. Cell. Biol. 267397-7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patrick, D. R., A. Oliff, and D. C. Heimbrook. 1994. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16 E7 protein. J. Biol. Chem. 2696842-6850. [PubMed] [Google Scholar]
- 27.Postlethwaite, R. 1970. Molluscum contagiosum. Arch. Environ. Health 21432-452. [DOI] [PubMed] [Google Scholar]
- 28.Rapoport, D. 2003. Finding the right organelle. EMBO Rep. 4948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogoff, H. A., M. T. Pickering, F. M. Frame, M. E. Debatis, Y. Sanchez, S. Jones, and T. F. Kowalik. 2004. Apoptosis associated with deregulated E2F activity is dependent on E2F1 and Atm/Nbs1/Chk2. Mol. Cell. Biol. 242968-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senkevich, T. G., J. J. Bugert, J. R. Sisler, E. V. Koonin, G. Darai, and B. Moss. 1996. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science 273813-816. [DOI] [PubMed] [Google Scholar]
- 31.Senkevich, T. G., E. V. Koonin, J. J. Bugert, G. Darai, and B. Moss. 1997. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 23319-42. [DOI] [PubMed] [Google Scholar]
- 32.Senkevich, T. G., and B. Moss. 1998. Domain structure, intracellular trafficking, and beta2-microglobulin binding of a major histocompatibility complex class I homolog encoded by molluscum contagiosum virus. Virology 250397-407. [DOI] [PubMed] [Google Scholar]
- 33.Shisler, J. L., T. G. Senkevich, M. J. Berry, and B. Moss. 1998. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science 279102-105. [DOI] [PubMed] [Google Scholar]
- 34.Shisler, J. L., and B. Moss. 2001. Molluscum contagiosum virus inhibitors of apoptosis: the MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology 28214-25. [DOI] [PubMed] [Google Scholar]
- 35.Simonart, T., I. Fayt, and J. C. Noel. 2002. An immunohistochemical study of abnormal keratinocyte proliferation in molluscum contagiosum. Br. J. Dermatol. 146609-614. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, T. L., S. T. Wasilenko, and M. Barry. 2005. Vaccinia virus F1L protein is a tail-anchored protein that functions at the mitochondria to inhibit apoptosis. J. Virol. 791084-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi, M., A. Izutani, and T. Tezuka. 1999. An immunohistochemical study of abnormal keratinocyte differentiation in molluscum contagiosum. Br. J. Dermatol. 141116-118. [DOI] [PubMed] [Google Scholar]
- 38.Verona, R., K. Moberg, S. Estes, M. Starz, J. P. Vernon, and J. A. Lees. 1997. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol. Cell. Biol. 177268-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wasilenko, S. T., T. L. Stewart, A. F. Meyers, and M. Barry. 2003. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. USA 10014345-14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81323-330. [DOI] [PubMed] [Google Scholar]
- 41.Whyte, P., N. M. Williamson, and E. Harlow. 1989. Cellular targets for transformation by the adenovirus E1A proteins. Cell 5667-75. [DOI] [PubMed] [Google Scholar]
- 42.Xiang, Y., and B. Moss. 1999. Identification of human and mouse homologs of the MC51L-53L-54L family of secreted glycoproteins encoded by the Molluscum contagiosum poxvirus. Virology 257297-302. [DOI] [PubMed] [Google Scholar]
- 43.Ziegler, H., W. Muranyi, H. G. Burgert, E. Kremmer, and U. H. Koszinowski. 2000. The luminal part of the murine cytomegalovirus glycoprotein gp40 catalyzes the retention of MHC class I molecules. EMBO J. 19870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]