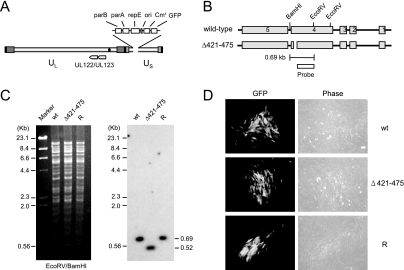
FIG. 2.
Generation of the recombinant T-BAC clone. (A) Genome structure of the T-BAC clone used in the present study. The F plasmid sequences containing the partition and replication functions (parA, parB, and repE), replication origin (ori), chloramphenicol resistance marker (Cmr), and the GFP eukaryotic expression cassette are indicated. (B) Genome structures of the T-BAC containing the wild-type or IE1(Δ421-475) gene. The region encompassing the MIE locus and the locations of the restriction enzyme sites used for mapping by Southern blot analysis are shown. The location for the 400-bp probe used for Southern blot analysis is shown. (C) Southern blot analysis. (Left) Ethidium bromide-stained total restriction DNA fragment patterns after EcoRV/BamHI digestion of three T-BAC DNAs (wild type [wt], Δ421-475, and revertant [R]) obtained by pulsed-field gel electrophoresis. (Right) Results of autoradiography after Southern blotting with 32P-labeled probe. Marker, λ-HindIII/EcoRI. (D) Infectivity of the transfected T-BAC DNAs in permissive HF cells. HF cells were electroporated with 2 μg of wild-type [wt], IE1(Δ421-475) mutant, or revertant [R] T-BAC DNAs. Each reaction also included 1 μg of plasmid pCMV71 encoding pp71 and 1 μg of plasmid pEGFP-C1. The cells were monitored for the spreading of the GFP signals. The GFP images and phase-contrast images were taken at 12 days after electroporation.