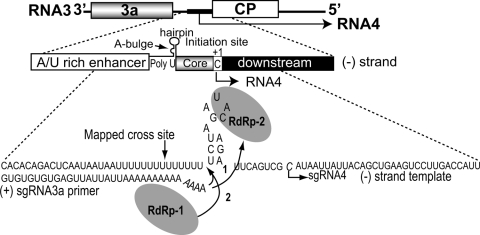
FIG. 4.
Two models of the crossing-over mechanism within the sgp region (27, 132). The top part shows the organization of sgp, while the sgp sequence is shown on the bottom. For clarity, only one recombining RNA3 molecule is shown, so the bowed arrows appear to depict crossovers toward sequences on the same template; however, more RNA3 templates might be engaged in the process. The ovals represent RdRp enzymes; one is leaving the template (RdRp-1), while another one (RdRp-2) is bound to the core hairpin of another (−)-strand template (for clarity, one template is shown). According to mechanism 1, the predetached 3′ poly(A) end “snatches” the RdRp-2 on another (−) strand, and the (+)-strand synthesis continues (represented by bow arrow 1). Mechanism 2 predicts that RdRp-1 detaches along with the sgRNA3a and rehybridizes to the poly(U) tract on another (−) strand to continue the (+)-strand synthesis (bowed arrow 2).