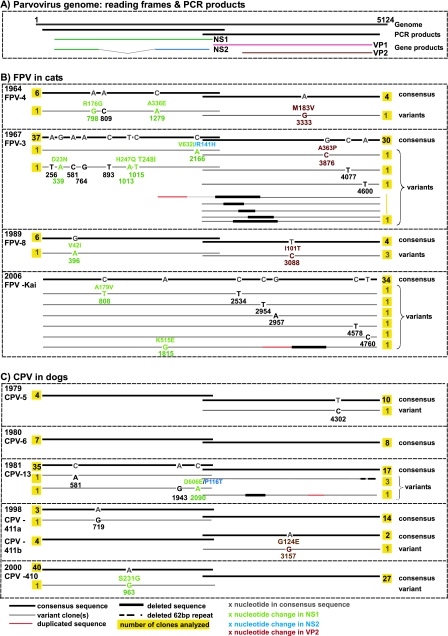
FIG. 3.
Intrahost diversity in FPV and CPV samples. (A) The gene regions covered by PCR amplification and a corresponding translation map of the parvovirus genome are indicated. (B and C) Divergent viral sequences detected in animals naturally infected with FPV (B) or CPV (C) are shown. The location of mutations in the parvovirus genome and the type of nucleotide substitution are indicated for each divergent sequence, the number of individual clones analyzed for each sample and the gene product affected by nonsynonymous mutations are indicated, and the year of isolation is identified. For comparison, in the case of the CPV-infected cat described previously by Battilani et al. (4), 10 out of 14 clones analyzed for the VP2 gene harbored one or more mutations each, which distinguished the clone from the consensus sequence.